Local bacteria affect the efficacy of chemotherapeutic drugs - PubMed (original) (raw)
Local bacteria affect the efficacy of chemotherapeutic drugs
Panos Lehouritis et al. Sci Rep. 2015.
Abstract
In this study, the potential effects of bacteria on the efficacy of frequently used chemotherapies was examined. Bacteria and cancer cell lines were examined in vitro and in vivo for changes in the efficacy of cancer cell killing mediated by chemotherapeutic agents. Of 30 drugs examined in vitro, the efficacy of 10 was found to be significantly inhibited by certain bacteria, while the same bacteria improved the efficacy of six others. HPLC and mass spectrometry analyses of sample drugs (gemcitabine, fludarabine, cladribine, CB1954) demonstrated modification of drug chemical structure. The chemoresistance or increased cytotoxicity observed in vitro with sample drugs (gemcitabine and CB1954) was replicated in in vivo murine subcutaneous tumour models. These findings suggest that bacterial presence in the body due to systemic or local infection may influence tumour responses or off-target toxicity during chemotherapy.
Figures
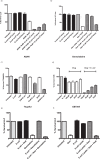
Figure 1. Tumour Cell Survival.
Cell survival assay. (a) E.coli at different cfu/ml were co-incubated with AQ4N (10 μM) after which the supernatant was applied directly to LLC cells (P < 0.01). (b) E. coli at different cfu/ml were co-incubated with gemcitabine (10 μM) after which the supernatant was directly applied to 4T1 Luc cells (P < 0.01). E.coli was co-incubated with Tegafur (c) or CB1954 (d) at the indicated concentrations after which the supernatant was directly applied to TRAMPC1 cells or CT26 (P < 0.01). Data (a–f) represent the average and standard error of four technical replicates. Data shown are representative of 3 independent experiments. (e) Tumour cell survival assay stained with MTS. Gemcitabine (10 μM) was incubated with live or heat killed E. coli (P < 0.001). (f) Tumour cell survival assay. Gemcitabine (10 μM) was incubated with either bacterial lysate (equivalent amounts to cell survival assay live bacteria dosages) alone or bacterial lysate that has been heat inactivated (P < 0.001). Data represent the average and standard error of four technical replicates. Data shown are representative of 2 independent experiments.
Figure 2. HPLC analysis of drug biotransformations.
Chromatograms: (a) Top; Vehicle alone (PBS DMSO 0.1%), Bottom; E. coli alone (b) Top; Gemcitabine, Bottom; Gemcitabine and E. coli. (c) Top; Fludarabine, Bottom; Fludarabine and E. coli (d) Top; Cladribine, Bottom; E. coli and Fludarabine. (e) Top; CB1954, Bottom; E. coli and CB1954 The drugs and their derivatives were detected by UV absorbance at 254 nm.
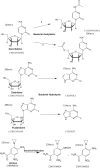
Figure 3. Schematic of drugs and proposed derivatives.
Structure prediction of drugs and derivatives based on mass spectrometry analysis. Hypothetical illustrations of structures based on elemental composition analysis and atomic mass fitting of HPLC peaks of drugs or drug derivatives after co-incubation with bacteria. For each molecule, its empirical formula and mass to charge ratio is also shown.
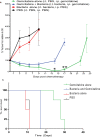
Figure 4. E. coli decreases the efficacy of gemcitabine in vivo.
Subcutaneous flank CT26 tumours growing in Balb/c mice were injected i.t with bacteria or PBS vehicle alone. Gemcitabine (60 mg/kg) was injected i.p. five times at three day intervals. (a) Tumour volume (%) relative to the first day of gemcitabine injection (day 0) is shown. *P < 0.03, **P = 0.002 (Bonferroni post hoc test) for gemcitabine alone versus gemcitabine + bacteria. (b) Kaplan-Meier plots showing mouse survival over time. The median survival post Day 0 of the gemcitabine + bacteria group was significantly less than that of the gemcitabine alone group (17 days vs. 28 days +/−1.25; P = 0.008). Data are expressed as mean ± SEM of 4 to 8 individual mice per group.
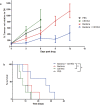
Figure 5. E. coli increases the cytotoxicity of CB1954.
Subcutaneous flank CT26 tumours growing in Balb/c mice were injected i.t with bacteria or PBS vehicle alone. CB1954 (20 mg/kg) was injected i.p. for the duration of the experiment at 3 day intervals. (a) Tumour volume (%) relative to the first day of CB1954 injection (day 0) is shown. (b) Kaplan-Meier plots showing mouse survival over time. The median survival post Day 0 of the CB1954 + bacteria group was significantly greater than that of the CB1954 alone group (26 days vs. 8 days. P = 0.0374). Data are expressed as mean ± SEM of 3-5 individual mice per group.
Similar articles
- Downregulation of deoxycytidine kinase in cytarabine-resistant mantle cell lymphoma cells confers cross-resistance to nucleoside analogs gemcitabine, fludarabine and cladribine, but not to other classes of anti-lymphoma agents.
Klanova M, Lorkova L, Vit O, Maswabi B, Molinsky J, Pospisilova J, Vockova P, Mavis C, Lateckova L, Kulvait V, Vejmelkova D, Jaksa R, Hernandez F, Trneny M, Vokurka M, Petrak J, Klener P Jr. Klanova M, et al. Mol Cancer. 2014 Jun 27;13:159. doi: 10.1186/1476-4598-13-159. Mol Cancer. 2014. PMID: 24972933 Free PMC article. - Modulation of cytarabine induced cytotoxicity using novel deoxynucleoside analogs in the HL60 cell line.
Hubeek I, Peters GJ, Broekhuizen AJ, Kaspers GJ. Hubeek I, et al. Nucleosides Nucleotides Nucleic Acids. 2004 Oct;23(8-9):1513-6. doi: 10.1081/NCN-200027727. Nucleosides Nucleotides Nucleic Acids. 2004. PMID: 15571288 - Phosphorylation of deoxycytidine kinase on Ser-74: impact on kinetic properties and nucleoside analog activation in cancer cells.
Amsailale R, Van Den Neste E, Arts A, Starczewska E, Bontemps F, Smal C. Amsailale R, et al. Biochem Pharmacol. 2012 Jul 1;84(1):43-51. doi: 10.1016/j.bcp.2012.03.022. Epub 2012 Apr 2. Biochem Pharmacol. 2012. PMID: 22490700 - [Clinical pharmacology of nucleoside analogues].
Milano G, Chamorey AL, Thyss A. Milano G, et al. Bull Cancer. 2002 Aug;89 Spec No:S71-5. Bull Cancer. 2002. PMID: 12449033 Review. French. - The role of membrane transporters in cellular resistance to anticancer nucleoside drugs.
Clarke ML, Mackey JR, Baldwin SA, Young JD, Cass CE. Clarke ML, et al. Cancer Treat Res. 2002;112:27-47. doi: 10.1007/978-1-4615-1173-1_2. Cancer Treat Res. 2002. PMID: 12481710 Review. No abstract available.
Cited by
- The effect of the intratumoral microbiome on tumor occurrence, progression, prognosis and treatment.
Gao F, Yu B, Rao B, Sun Y, Yu J, Wang D, Cui G, Ren Z. Gao F, et al. Front Immunol. 2022 Nov 18;13:1051987. doi: 10.3389/fimmu.2022.1051987. eCollection 2022. Front Immunol. 2022. PMID: 36466871 Free PMC article. Review. - Metabolic Phenotyping Predicts Gemcitabine and Cisplatin Chemosensitivity in Patients With Cholangiocarcinoma.
Suksawat M, Phetcharaburanin J, Klanrit P, Namwat N, Khuntikeo N, Titapun A, Jarearnrat A, Vilayhong V, Sa-Ngiamwibool P, Techasen A, Wangwiwatsin A, Mahalapbutr P, Li JV, Loilome W. Suksawat M, et al. Front Public Health. 2022 Feb 10;10:766023. doi: 10.3389/fpubh.2022.766023. eCollection 2022. Front Public Health. 2022. PMID: 35223723 Free PMC article. - Intratumoral Fusobacterium Nucleatum Levels Predict Therapeutic Response to Neoadjuvant Chemotherapy in Esophageal Squamous Cell Carcinoma.
Yamamura K, Izumi D, Kandimalla R, Sonohara F, Baba Y, Yoshida N, Kodera Y, Baba H, Goel A. Yamamura K, et al. Clin Cancer Res. 2019 Oct 15;25(20):6170-6179. doi: 10.1158/1078-0432.CCR-19-0318. Epub 2019 Jul 29. Clin Cancer Res. 2019. PMID: 31358543 Free PMC article. - The gut microbiome: an orchestrator of xenobiotic metabolism.
Collins SL, Patterson AD. Collins SL, et al. Acta Pharm Sin B. 2020 Jan;10(1):19-32. doi: 10.1016/j.apsb.2019.12.001. Epub 2019 Dec 10. Acta Pharm Sin B. 2020. PMID: 31998605 Free PMC article. Review. - Stress and stability: applying the Anna Karenina principle to animal microbiomes.
Zaneveld JR, McMinds R, Vega Thurber R. Zaneveld JR, et al. Nat Microbiol. 2017 Aug 24;2:17121. doi: 10.1038/nmicrobiol.2017.121. Nat Microbiol. 2017. PMID: 28836573
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources