Chromatin dynamics after DNA damage: The legacy of the access-repair-restore model - PubMed (original) (raw)
Review
Chromatin dynamics after DNA damage: The legacy of the access-repair-restore model
Sophie E Polo et al. DNA Repair (Amst). 2015 Dec.
Abstract
Eukaryotic genomes are packaged into chromatin, which is the physiological substrate for all DNA transactions, including DNA damage and repair. Chromatin organization imposes major constraints on DNA damage repair and thus undergoes critical rearrangements during the repair process. These rearrangements have been integrated into the "access-repair-restore" (ARR) model, which provides a molecular framework for chromatin dynamics in response to DNA damage. Here, we take a historical perspective on the elaboration of this model and describe the molecular players involved in damaged chromatin reorganization in human cells. In particular, we present our current knowledge of chromatin assembly coupled to DNA damage repair, focusing on the role of histone variants and their dedicated chaperones. Finally, we discuss the impact of chromatin rearrangements after DNA damage on chromatin function and epigenome maintenance.
Keywords: Chromatin assembly; DNA damage repair; Epigenome maintenance; Histone chaperones; Histone variants.
Copyright © 2015 Elsevier B.V. All rights reserved.
Conflict of interest statement
statement None
Figures
Fig.1. The Access-Repair-Restore model: from the initial concepts to the most recent molecular principles
Nucleosome rearrangements during repair of damaged chromatin in mammalian cells as described in the Unfolding-Refolding model (A), which developed in the Unfolding-Refolding-Repositioning model (B), Access-Repair-Restore model (C) and, more recently, in the Access/Prime-Repair/Restore model (D). DNA is represented in purple, the repair synthesis patch in pink, histone modifications in orange, inner core histones in grey, outer core histones in dark blue and newly synthesized histones in green.
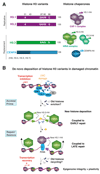
Fig.2. Restoration of UVC-damaged chromatin structure and function through de novo deposition of H3 variants
A. Main histone H3 variants and their dedicated chaperones in mammalian cells Amino acids that differ between the histone variant sequences are indicated except for the more divergent centromeric variant CENPA. Less well characterized H3 variants are listed in parentheses. CAF1, HIRA, HJURP are histone chaperones. B. Restoration of chromatin organization after UVC damage involves de novo deposition of H3 variants Repair factors that facilitate the recruitment of the histone chaperones HIRA and CAF-1 to UV-damaged chromatin regions are depicted in blue. HIRA-mediated deposition of new H3.3 (green) precedes CAF-1-dependent incorporation of new H3.1 (purple). Early bookmarking of chromatin by the H3.3 chaperone HIRA is required for restoring transcriptional activity (red) after completion of DNA repair. While new histone deposition in UVC damaged chromatin is firmly established, the dynamics of old histones and their contribution to repaired chromatin are still to be determined. Adapted from [81] with permission.
Similar articles
- Reshaping chromatin after DNA damage: the choreography of histone proteins.
Polo SE. Polo SE. J Mol Biol. 2015 Feb 13;427(3):626-36. doi: 10.1016/j.jmb.2014.05.025. Epub 2014 Jun 2. J Mol Biol. 2015. PMID: 24887097 Free PMC article. Review. - Chromatin dynamics during nucleotide excision repair: histones on the move.
Adam S, Polo SE. Adam S, et al. Int J Mol Sci. 2012;13(9):11895-11911. doi: 10.3390/ijms130911895. Epub 2012 Sep 19. Int J Mol Sci. 2012. PMID: 23109890 Free PMC article. Review. - Chromatin plasticity in response to DNA damage: The shape of things to come.
Adam S, Dabin J, Polo SE. Adam S, et al. DNA Repair (Amst). 2015 Aug;32:120-126. doi: 10.1016/j.dnarep.2015.04.022. Epub 2015 May 1. DNA Repair (Amst). 2015. PMID: 25957486 Free PMC article. Review. - Chromatin dynamics and DNA replication roadblocks.
Hammond-Martel I, Verreault A, Wurtele H. Hammond-Martel I, et al. DNA Repair (Amst). 2021 Aug;104:103140. doi: 10.1016/j.dnarep.2021.103140. Epub 2021 May 23. DNA Repair (Amst). 2021. PMID: 34087728 Review.
Cited by
- Damage sensor role of UV-DDB during base excision repair.
Jang S, Kumar N, Beckwitt EC, Kong M, Fouquerel E, Rapić-Otrin V, Prasad R, Watkins SC, Khuu C, Majumdar C, David SS, Wilson SH, Bruchez MP, Opresko PL, Van Houten B. Jang S, et al. Nat Struct Mol Biol. 2019 Aug;26(8):695-703. doi: 10.1038/s41594-019-0261-7. Epub 2019 Jul 22. Nat Struct Mol Biol. 2019. PMID: 31332353 Free PMC article. - Nucleosome remodeling at origins of global genome-nucleotide excision repair occurs at the boundaries of higher-order chromatin structure.
van Eijk P, Nandi SP, Yu S, Bennett M, Leadbitter M, Teng Y, Reed SH. van Eijk P, et al. Genome Res. 2019 Jan;29(1):74-84. doi: 10.1101/gr.237198.118. Epub 2018 Dec 14. Genome Res. 2019. PMID: 30552104 Free PMC article. - Chaperoning histones at the DNA repair dance.
Chakraborty U, Shen ZJ, Tyler J. Chakraborty U, et al. DNA Repair (Amst). 2021 Dec;108:103240. doi: 10.1016/j.dnarep.2021.103240. Epub 2021 Oct 13. DNA Repair (Amst). 2021. PMID: 34687987 Free PMC article. Review. - Histone H1 acetylation at lysine 85 regulates chromatin condensation and genome stability upon DNA damage.
Li Y, Li Z, Dong L, Tang M, Zhang P, Zhang C, Cao Z, Zhu Q, Chen Y, Wang H, Wang T, Lv D, Wang L, Zhao Y, Yang Y, Wang H, Zhang H, Roeder RG, Zhu WG. Li Y, et al. Nucleic Acids Res. 2018 Sep 6;46(15):7716-7730. doi: 10.1093/nar/gky568. Nucleic Acids Res. 2018. PMID: 29982688 Free PMC article. - Human CRL4DDB2 ubiquitin ligase preferentially regulates post-repair chromatin restoration of H3K56Ac through recruitment of histone chaperon CAF-1.
Zhu Q, Wei S, Sharma N, Wani G, He J, Wani AA. Zhu Q, et al. Oncotarget. 2017 Oct 17;8(61):104525-104542. doi: 10.18632/oncotarget.21869. eCollection 2017 Nov 28. Oncotarget. 2017. PMID: 29262658 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources