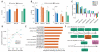Somatic mutation in single human neurons tracks developmental and transcriptional history - PubMed (original) (raw)
. 2015 Oct 2;350(6256):94-98.
doi: 10.1126/science.aab1785.
Mollie B Woodworth # 1, Semin Lee # 2, Gilad D Evrony 1, Bhaven K Mehta 1, Amir Karger 3, Soohyun Lee 2, Thomas W Chittenden 3 4, Alissa M D'Gama 1, Xuyu Cai 1, Lovelace J Luquette 2, Eunjung Lee 2 5, Peter J Park 2 5, Christopher A Walsh 1
Affiliations
- PMID: 26430121
- PMCID: PMC4664477
- DOI: 10.1126/science.aab1785
Somatic mutation in single human neurons tracks developmental and transcriptional history
Michael A Lodato et al. Science. 2015.
Abstract
Neurons live for decades in a postmitotic state, their genomes susceptible to DNA damage. Here we survey the landscape of somatic single-nucleotide variants (SNVs) in the human brain. We identified thousands of somatic SNVs by single-cell sequencing of 36 neurons from the cerebral cortex of three normal individuals. Unlike germline and cancer SNVs, which are often caused by errors in DNA replication, neuronal mutations appear to reflect damage during active transcription. Somatic mutations create nested lineage trees, allowing them to be dated relative to developmental landmarks and revealing a polyclonal architecture of the human cerebral cortex. Thus, somatic mutations in the brain represent a durable and ongoing record of neuronal life history, from development through postmitotic function.
Copyright © 2015, American Association for the Advancement of Science.
Figures
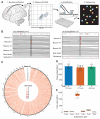
Fig. 1. Somatic SNVs are detected by single-neuron whole-genome sequencing
(A) Schematic of approach. Nuclei are isolated from a 0.5-cm3 piece of frozen postmortem tissue by fluorescence-activated nuclear sorting (FANS), and DNA is amplified by Φ29 polymerase-mediated multiple-displacement amplification (MDA) and subjected to whole-genome sequencing (WGS). (B) Sample sequencing alignment tracks from brain B. One SNV is called uniquely in one sample (left), and one is shared between neurons 2 and 77 but absent from other single cells and heart (right). (C) Circos plot of SNV rate per megabase in brain B across human autosomes demonstrates that somatic mutations are distributed broadly. (D) SNVs per neuron are tightly correlated within each individual. (E) Most single-neuron SNVs in brain B are C>T transitions. Error bars in (D) and (E) denote SD; data points from individual neurons are spread horizontally for visibility.
Fig. 2. Somatic SNVs occur at loci that are expressed in the brain and associated with nervous system function and disease
(A) Coding exons are enriched for somatic SNVs. *P < 0.05, combined binomial test, Bonferroni-corrected. (B) SNVs in transcribed regions display a strand bias, suggesting that transcriptional damage influences the somatic SNV rate. *P < 0.05, analysis of variance with Sidak’s correction for multiple testing. Error bars in (A) and (B) denote SD. (C) Single-neuron SNVs correlate with epigenetic marks of transcription in the fetal brain and are depleted in heterochromatin, in opposition to the pattern observed in glioblastoma multiforme (GBM) SNVs. (D) Single-neuron SNVs occur in genes expressed in the cerebral cortex. Genes in the lowest expression quartile were significantly depleted, and those in the third quartile were significantly enriched, for single-neuron SNVs. *P < 0.05, +P < 0.05, Fisher combined one-tailed Poisson P value for enrichment and depletion of SNVs, respectively. (E) Gene ontology categories associated with nervous system development and function are enriched for mutated genes across single neurons from brain B. (F) Examples of genes implicated in human disease that were mutated in single brain B neurons (for full list, see table S8). Green SNVs occurred in introns or downstream of the coding region, the orange SNV occurred in an exon and induced a missense mutation, and the magenta SNV is a nonsense mutation.
Fig. 3. Somatic mutations are shared between multiple neurons and demonstrate lineage relationships
(A) Lineage map of 136 human cortical neurons from brain B derived from 18 clonal somatic mutations, including SNVs, long interspersed nuclear element (LINE) insertions, and a TG-dinucleotide expansion. Neurons are placed into four distinct nested clades (pink, green, blue, purple) defined by one or more independent mutations. Cells are ordered within clades according to the presence of multiple somatic mutations. A few cells in each clade fail to manifest individual SNVs shared by other cells of the same clade (indicated by open squares), likely representing incomplete amplification (fig. S2). Dark gray boxes represent cells analyzed by WGS; light gray represents cells analyzed by Sanger-based genotyping. Genomic locations of somatic mutations are given in fig. S11. (B) Ultradeep sequencing of mutated loci across the cortex of brain B. Clonal SNVs from a single clade are progressively regionally restricted to frontal cortex and become progressively rarer in bulk tissue, reflecting their later origin during development and neurogenesis. Blue circle, mutation present; empty circle, mutation absent; blue shading, likely spatial distribution of mutation. Percentage range of heterozygous cells is indicated for each SNV. (C) Ultradeep sequencing of mutated loci across the brain and body. Some variants are brain-specific (top) and others are shared across germ layers (bottom). Samples sequenced are prefrontal cortex [Brodmann area (BA) 10/BA46], cingulate cortex (BA32/BA8), temporal cortex (BA38), cerebellum (Cb), spinal cord (SC), aorta (Ao), heart (He), liver (Li), lung (Lu), and pancreas (Pa). (D) Genotyping shared variants in small sections of human cortex. Left: 4′,6-diamidino-2-phenylindole (DAPI) stain of segment of representative section; scale bar, 200 μm. Center: Three consecutive 300-μm coronal sections from BA40 (red, upper left) were dissected into three axial regions each (1 to 9). Right: Genotyping results for dissected sections. Solid circles denote presence of mutation in indicated sample; open circles denote absence. Mutations with high allele fractions are present in all or virtually all regions, whereas only the least prevalent somatic variant (present in <0.5% of cells) is present in one region but not most regions.
Comment in
- NEUROSCIENCE. A tree of the human brain.
Linnarsson S. Linnarsson S. Science. 2015 Oct 2;350(6256):37. doi: 10.1126/science.aad2792. Science. 2015. PMID: 26430106 No abstract available.
Similar articles
- NEUROSCIENCE. A tree of the human brain.
Linnarsson S. Linnarsson S. Science. 2015 Oct 2;350(6256):37. doi: 10.1126/science.aad2792. Science. 2015. PMID: 26430106 No abstract available. - Cell lineage analysis in human brain using endogenous retroelements.
Evrony GD, Lee E, Mehta BK, Benjamini Y, Johnson RM, Cai X, Yang L, Haseley P, Lehmann HS, Park PJ, Walsh CA. Evrony GD, et al. Neuron. 2015 Jan 7;85(1):49-59. doi: 10.1016/j.neuron.2014.12.028. Neuron. 2015. PMID: 25569347 Free PMC article. - Parallel RNA and DNA analysis after deep sequencing (PRDD-seq) reveals cell type-specific lineage patterns in human brain.
Huang AY, Li P, Rodin RE, Kim SN, Dou Y, Kenny CJ, Akula SK, Hodge RD, Bakken TE, Miller JA, Lein ES, Park PJ, Lee EA, Walsh CA. Huang AY, et al. Proc Natl Acad Sci U S A. 2020 Jun 23;117(25):13886-13895. doi: 10.1073/pnas.2006163117. Epub 2020 Jun 10. Proc Natl Acad Sci U S A. 2020. PMID: 32522880 Free PMC article. - Intersection of diverse neuronal genomes and neuropsychiatric disease: The Brain Somatic Mosaicism Network.
McConnell MJ, Moran JV, Abyzov A, Akbarian S, Bae T, Cortes-Ciriano I, Erwin JA, Fasching L, Flasch DA, Freed D, Ganz J, Jaffe AE, Kwan KY, Kwon M, Lodato MA, Mills RE, Paquola ACM, Rodin RE, Rosenbluh C, Sestan N, Sherman MA, Shin JH, Song S, Straub RE, Thorpe J, Weinberger DR, Urban AE, Zhou B, Gage FH, Lehner T, Senthil G, Walsh CA, Chess A, Courchesne E, Gleeson JG, Kidd JM, Park PJ, Pevsner J, Vaccarino FM; Brain Somatic Mosaicism Network. McConnell MJ, et al. Science. 2017 Apr 28;356(6336):eaal1641. doi: 10.1126/science.aal1641. Epub 2017 Apr 27. Science. 2017. PMID: 28450582 Free PMC article. Review. - Why Cortical Neurons Cannot Divide, and Why Do They Usually Die in the Attempt?
Aranda-Anzaldo A, Dent MA. Aranda-Anzaldo A, et al. J Neurosci Res. 2017 Apr;95(4):921-929. doi: 10.1002/jnr.23765. Epub 2016 Jul 12. J Neurosci Res. 2017. PMID: 27402311 Review.
Cited by
- Applications of Single-Cell DNA Sequencing.
Evrony GD, Hinch AG, Luo C. Evrony GD, et al. Annu Rev Genomics Hum Genet. 2021 Aug 31;22:171-197. doi: 10.1146/annurev-genom-111320-090436. Epub 2021 Mar 15. Annu Rev Genomics Hum Genet. 2021. PMID: 33722077 Free PMC article. Review. - Decoding and recoding plant development.
Guiziou S, Chu JC, Nemhauser JL. Guiziou S, et al. Plant Physiol. 2021 Oct 5;187(2):515-526. doi: 10.1093/plphys/kiab336. Plant Physiol. 2021. PMID: 35237818 Free PMC article. Review. - Human Brain Single Nucleotide Polymorphism: Validation of DNA Sequencing.
Picher ÁJ, Hernández F, Budeus B, Soriano E, Avila J. Picher ÁJ, et al. J Alzheimers Dis Rep. 2018 May 31;2(1):103-109. doi: 10.3233/ADR-170039. J Alzheimers Dis Rep. 2018. PMID: 30480253 Free PMC article. Review. - Heteroplasmic mitochondrial DNA mutations in frontotemporal lobar degeneration.
Nie Y, Murley A, Golder Z, Rowe JB, Allinson K, Chinnery PF. Nie Y, et al. Acta Neuropathol. 2022 Jun;143(6):687-695. doi: 10.1007/s00401-022-02423-6. Epub 2022 Apr 30. Acta Neuropathol. 2022. PMID: 35488929 Free PMC article. - LINE-1 regulates cortical development by acting as long non-coding RNAs.
Mangoni D, Simi A, Lau P, Armaos A, Ansaloni F, Codino A, Damiani D, Floreani L, Di Carlo V, Vozzi D, Persichetti F, Santoro C, Pandolfini L, Tartaglia GG, Sanges R, Gustincich S. Mangoni D, et al. Nat Commun. 2023 Aug 17;14(1):4974. doi: 10.1038/s41467-023-40743-7. Nat Commun. 2023. PMID: 37591988 Free PMC article.
References
- De S. Trends Genet. 2011;27:217–223. - PubMed
- Dumanski JP, Piotrowski A. Methods Mol. Biol. 2011;838:249–272. - PubMed
Publication types
MeSH terms
Grants and funding
- U01 MH106883/MH/NIMH NIH HHS/United States
- T32 GM007226/GM/NIGMS NIH HHS/United States
- T32 AG000222/AG/NIA NIH HHS/United States
- 1S10RR028832-01/RR/NCRR NIH HHS/United States
- T32 GM007753/GM/NIGMS NIH HHS/United States
- R01 NS079277/NS/NINDS NIH HHS/United States
- P50 MH106933/MH/NIMH NIH HHS/United States
- HHMI/Howard Hughes Medical Institute/United States
- S10 RR028832/RR/NCRR NIH HHS/United States
- R01 NS032457/NS/NINDS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources