Single Cell RNA-Sequencing of Pluripotent States Unlocks Modular Transcriptional Variation - PubMed (original) (raw)
. 2015 Oct 1;17(4):471-85.
doi: 10.1016/j.stem.2015.09.011.
Jong Kyoung Kim 2, Jason C H Tsang 3, Tomislav Ilicic 1, Johan Henriksson 2, Kedar N Natarajan 1, Alex C Tuck 4, Xuefei Gao 3, Marc Bühler 5, Pentao Liu 3, John C Marioni 6, Sarah A Teichmann 7
Affiliations
- PMID: 26431182
- PMCID: PMC4595712
- DOI: 10.1016/j.stem.2015.09.011
Single Cell RNA-Sequencing of Pluripotent States Unlocks Modular Transcriptional Variation
Aleksandra A Kolodziejczyk et al. Cell Stem Cell. 2015.
Abstract
Embryonic stem cell (ESC) culture conditions are important for maintaining long-term self-renewal, and they influence cellular pluripotency state. Here, we report single cell RNA-sequencing of mESCs cultured in three different conditions: serum, 2i, and the alternative ground state a2i. We find that the cellular transcriptomes of cells grown in these conditions are distinct, with 2i being the most similar to blastocyst cells and including a subpopulation resembling the two-cell embryo state. Overall levels of intercellular gene expression heterogeneity are comparable across the three conditions. However, this masks variable expression of pluripotency genes in serum cells and homogeneous expression in 2i and a2i cells. Additionally, genes related to the cell cycle are more variably expressed in the 2i and a2i conditions. Mining of our dataset for correlations in gene expression allowed us to identify additional components of the pluripotency network, including Ptma and Zfp640, illustrating its value as a resource for future discovery.
Copyright © 2015 The Authors. Published by Elsevier Inc. All rights reserved.
Figures
Graphical abstract
Figure 1
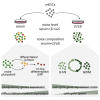
Experimental Scheme of Hybrid mESCs in Three Culture Conditions Schematic of experimental setup and cell culture conditions used in our study.
Figure 2
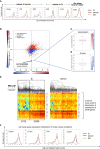
Global Cell-to-Cell Variation in Gene Expression (A) Gene expression distributions of genes, which are noisier in 2i than serum, that have similar noise profiles in serum (red), 2i (blue), and a2i (yellow). Distributions of gene expression were smoothed using the kernel density estimation function in R with default parameters. Tcerg1 does not have significantly different expression profiles between culture conditions (two-sided KS test p value for 2i and a2i comparison is 0.82, and for 2i and serum, 0.16). By contrast, other genes, such as Ccnb1, are more heterogeneous in 2i (p = 7 × 10−4 by two-sided KS test between 2i and serum), while some, such as Nanog, Klf4, or Nr0b1, are more heterogeneous in serum (p < 10−15 by two-sided KS test between 2i and serum for genes shown). (B) Comparison of the levels of gene expression and noise for gene ontology (GO) categories between serum and 2i (excluding 2i replicates containing 2C-like cells). The logarithm (log10) of p values from two-sided paired t tests applied to mean normalized read counts (x axis) and DMs (y axis) was computed for each GO category and plotted against each other by multiplying the sign of the t statistic. (C and D) Example of a GO category (GO:0000280, nuclear division) that is noisier in 2i (C) and is similarly expressed between the two conditions (D). (E) Heatmaps showing the expression of cell-cycle-related genes in serum and 2i, with a distinct separation into G1/S versus G2/M cells in 2i, with less distinction between individual cells in serum. (F) Gene expression profiles for key cell-cycle genes in all conditions show more heterogeneity in 2i.
Figure 3
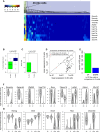
Population Structure in Serum, 2i, and a2i Cells (A) Clustering of cells in three culture conditions using a panel of pluripotency factors and differentiation markers. Correlations between cells and genes were calculated using Spearman correlation. Below the heatmap we show a model of the subpopulations of cells grown in serum. The schematic shows cells that express differentiation markers (red), cells that are primed for differentiation while remaining pluripotent (orange), and cells that are closest to the ground state of pluripotency (green). (B and C) Gene expression distributions of genes that become downregulated (B) and upregulated (C) upon differentiation. Expression is shown as log2 size factor normalized counts. Oct4 expression is similar in cells closer to the ground state of pluripotency (green) and cells that are primed for differentiation (yellow), and it is much lower in cells we defined as moving toward differentiation (red). (D) Gene expression distributions of cell-cycle genes.
Figure 4
Clustering of mESCs Grown in Serum, 2i, and a2i Media (A) All cells (n = 704) grown in the three different culture conditions are projected onto the first two principal components. All genes with mean normalized read counts larger than ten were considered and principal component analysis (PCA) was performed. (B) Distribution of genes contributing to PC1. (C) GO enrichment analysis of genes most strongly contributing to PC1 separation. (D) PCA loading plot of the Spearman’s rank correlation coefficients from mESCs and single cells of mouse preimplantation embryos (Deng et al., 2014) showing the mapping of mESCs in mouse development stages. The cells are visualized by loadings of the first three principal components of the Spearman’s rank correlation matrix between cells, where we used the same expression cutoff as that employed by Deng et al. (E) PCA of Spearman’s rank correlation matrix between cells from three conditions and blastocyst. The first three components are shown.
Figure 5
2C-like Population (A) Clustering of cells grown in 2i using markers of the 2C-like state (Macfarlan et al., 2012). Correlations were calculated using Spearman correlation. The dendrogram divides cells into two groups, one of which contains ten cells expressing 2C-markers. (B) Boxplot showing percentage of reads mapping to the exons in both subpopulations of cells in 2i. p was calculated using a Wilcoxon test. (C) Boxplot showing RPM (reads per million) mapping to the MERVL retrovirus in both subpopulations of cells in 2i. p was calculated using a Wilcoxon test. (D) Mean expression of genes reported to be at least 2-fold upregulated or downregulated in 2C-like cells (Macfarlan et al., 2012) in cells that we identified as 2C-like cells and in the remaining 2i cells. (E) Barplot showing the number of significantly (DESeq, adjusted p < 0.05) upregulated and downregulated genes in 2C-like cells. (F) Gene expression distributions of genes that become upregulated or downregulated in 2C-like cells (2C) in comparison to remaining cells grown in 2i media (2i). (G) Expression of key pluripotency genes in 2C-like cells (2C), the rest of cells grown in 2i media (2i), cells from the two-cell stage (2cell), and cells from the blastocyst stage (blast) of the embryo.
Figure 6
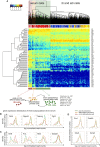
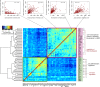
Spearman Correlation Matrix of Transcription Factors and Key Pluripotency Genes The heatmap shows the correlation coefficients between a set of transcription factors and other key genes involved in pluripotency. Above are examples of genes with expression patterns that correlate positively and negatively (from the left: Zfp42 and Creb3, Zfp42 and Nanog, Tet1 and Tet2, Tet1 and Jarid2).
Figure 7
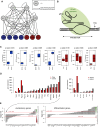
Validation of Putative Members of the Pluripotency Network (A) Network showing known interactions of core pluripotency factors with the novel candidate genes. Data obtained from ChIP-seq and ChIP-ChIP experiments from ESCAPE database. (B) Schematic showing experimental design. Catalytically inactive Cas9 and gRNA bind to the promoter of the targeted gene, occluding it and competing for binding with transcription factors and polymerases. (C) Expression level of repressed genes in samples and control. Targets with significant repression are in blue. (D) Barplot of gene expression levels of significantly differentially expressed genes in Ptma- and Zfp640-repressed samples (DESeq, multiple hypotheses testing adjusted p < 0.05). (E) Barplots showing the logarithm of p values for differential expression from DESeq of pluripotency (left) and differentiation (right) genes in the Ptma knockdown samples. For genes that are downregulated, the numbers are negative, and they are positive for upregulated genes. The red line indicates a p threshold of 0.05.
Comment in
- Moving Toward the Ground State.
Kumar I, Ivanova N. Kumar I, et al. Cell Stem Cell. 2015 Oct 1;17(4):375-6. doi: 10.1016/j.stem.2015.09.009. Cell Stem Cell. 2015. PMID: 26431178
Similar articles
- Low Focal Adhesion Signaling Promotes Ground State Pluripotency of Mouse Embryonic Stem Cells.
Taleahmad S, Mirzaei M, Samadian A, Hassani SN, Haynes PA, Salekdeh GH, Baharvand H. Taleahmad S, et al. J Proteome Res. 2017 Oct 6;16(10):3585-3595. doi: 10.1021/acs.jproteome.7b00322. Epub 2017 Sep 13. J Proteome Res. 2017. PMID: 28850235 - 2i Maintains a Naive Ground State in ESCs through Two Distinct Epigenetic Mechanisms.
Sim YJ, Kim MS, Nayfeh A, Yun YJ, Kim SJ, Park KT, Kim CH, Kim KS. Sim YJ, et al. Stem Cell Reports. 2017 May 9;8(5):1312-1328. doi: 10.1016/j.stemcr.2017.04.001. Epub 2017 Apr 27. Stem Cell Reports. 2017. PMID: 28457889 Free PMC article. - Ground State Conditions Induce Rapid Reorganization of Core Pluripotency Factor Binding before Global Epigenetic Reprogramming.
Galonska C, Ziller MJ, Karnik R, Meissner A. Galonska C, et al. Cell Stem Cell. 2015 Oct 1;17(4):462-70. doi: 10.1016/j.stem.2015.07.005. Epub 2015 Jul 30. Cell Stem Cell. 2015. PMID: 26235340 Free PMC article. - Open Chromatin, Epigenetic Plasticity, and Nuclear Organization in Pluripotency.
Schlesinger S, Meshorer E. Schlesinger S, et al. Dev Cell. 2019 Jan 28;48(2):135-150. doi: 10.1016/j.devcel.2019.01.003. Dev Cell. 2019. PMID: 30695696 Review. - Transcription regulation and chromatin structure in the pluripotent ground state.
Marks H, Stunnenberg HG. Marks H, et al. Biochim Biophys Acta. 2014 Mar;1839(3):129-37. doi: 10.1016/j.bbagrm.2013.09.005. Epub 2013 Oct 2. Biochim Biophys Acta. 2014. PMID: 24096207 Review.
Cited by
- Identifying cell types by lasso-constraint regularized Gaussian graphical model based on weighted distance penalty.
Zhang W, Xu Y, Zheng X, Shen J, Li Y. Zhang W, et al. Brief Bioinform. 2024 Sep 23;25(6):bbae572. doi: 10.1093/bib/bbae572. Brief Bioinform. 2024. PMID: 39541187 Free PMC article. - DNA methylation shapes the Polycomb landscape during the exit from naive pluripotency.
Richard Albert J, Urli T, Monteagudo-Sánchez A, Le Breton A, Sultanova A, David A, Scarpa M, Schulz M, Greenberg MVC. Richard Albert J, et al. Nat Struct Mol Biol. 2024 Oct 24. doi: 10.1038/s41594-024-01405-4. Online ahead of print. Nat Struct Mol Biol. 2024. PMID: 39448850 - Clustering scRNA-seq data with the cross-view collaborative information fusion strategy.
Lou Z, Wei X, Hu Y, Hu S, Wu Y, Tian Z. Lou Z, et al. Brief Bioinform. 2024 Sep 23;25(6):bbae511. doi: 10.1093/bib/bbae511. Brief Bioinform. 2024. PMID: 39402696 Free PMC article. - Evaluating Strategies to Assess the Differentiation Potential of Human Pluripotent Stem Cells: A Review, Analysis and Call for Innovation.
Smith L, Quelch-Cliffe R, Liu F, Aguilar AH, Przyborski S. Smith L, et al. Stem Cell Rev Rep. 2024 Sep 28. doi: 10.1007/s12015-024-10793-5. Online ahead of print. Stem Cell Rev Rep. 2024. PMID: 39340737 Review. - scVGATAE: A Variational Graph Attentional Autoencoder Model for Clustering Single-Cell RNA-seq Data.
Liu L, Wu X, Yu J, Zhang Y, Niu K, Yu A. Liu L, et al. Biology (Basel). 2024 Sep 11;13(9):713. doi: 10.3390/biology13090713. Biology (Basel). 2024. PMID: 39336140 Free PMC article.
References
- Bibel M., Richter J., Lacroix E., Barde Y.A. Generation of a defined and uniform population of CNS progenitors and neurons from mouse embryonic stem cells. Nat. Protoc. 2007;2:1034–1043. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- WT_/Wellcome Trust/United Kingdom
- 22231/CRUK_/Cancer Research UK/United Kingdom
- 103977/WT_/Wellcome Trust/United Kingdom
- BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources