Thermal Degradation of Small Molecules: A Global Metabolomic Investigation - PubMed (original) (raw)
Thermal Degradation of Small Molecules: A Global Metabolomic Investigation
Mingliang Fang et al. Anal Chem. 2015.
Abstract
Thermal processes are widely used in small molecule chemical analysis and metabolomics for derivatization, vaporization, chromatography, and ionization, especially in gas chromatography mass spectrometry (GC/MS). In this study the effect of heating was examined on a set of 64 small molecule standards and, separately, on human plasma metabolite extracts. The samples, either derivatized or underivatized, were heated at three different temperatures (60, 100, and 250 °C) at different exposure times (30 s, 60 s, and 300 s). All the samples were analyzed by liquid chromatography coupled to electrospray ionization mass spectrometry (LC/MS) and the data processed by XCMS Online ( xcmsonline.scripps.edu ). The results showed that heating at an elevated temperature of 100 °C had an appreciable effect on both the underivatized and derivatized molecules, and heating at 250 °C created substantial changes in the profile. For example, over 40% of the molecular peaks were altered in the plasma metabolite analysis after heating (250 °C, 300s) with a significant formation of degradation and transformation products. The analysis of 64 small molecule standards validated the temperature-induced changes observed on the plasma metabolites, where most of the small molecules degraded at elevated temperatures even after minimal exposure times (30 s). For example, tri- and diorganophosphates (e.g., adenosine triphosphate and adenosine diphosphate) were readily degraded into a mono-organophosphate (e.g., adenosine monophosphate) during heating. Nucleosides and nucleotides (e.g., inosine and inosine monophosphate) were also found to be transformed into purine derivatives (e.g., hypoxanthine). A newly formed transformation product, oleoyl ethyl amide, was identified in both the underivatized and derivatized forms of the plasma extracts and small molecule standard mixture, and was likely generated from oleic acid. Overall these analyses show that small molecules and metabolites undergo significant time-sensitive alterations when exposed to elevated temperatures, especially those conditions that mimic sample preparation and analysis in GC/MS experiments.
Figures
Figure 1
Global molecular profiling approach used to investigate heating on small molecule stability.
Figure 2
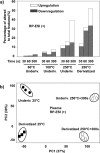
Percentage of altered (both upregulated and downregulated) features compared to the total features in the underivatized and derivatized plasma small molecule metabolites heated at 60 °C, 100 °C, and 250 °C for 30 s, 60 s, and 300 s. The subsequent analyses were performed using (a) RP-ESI (+) and overall results displayed with (b) principal component analysis scores plots of 25 and 250 °C for 300 s heated metabolites. All the upregulated and downregulated features were from pairwise comparison of each tested condition with room temperature (25 °C) and filtered using p < 0.01.
Figure 3
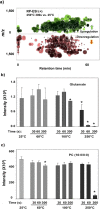
(a) Cloud plot showing thousands of altered features (represented by “bubbles”) from the underivatized plasma metabolites heated at 250 °C for 300 s (filtered by fold change >1.5 and p < 0.01). (b and c) Response change of glutamate and PC(16:0/0:0) heated at different temperatures and durations. The results and the standard deviation were obtained from triplicate analysis. “*” represent the samples which are significantly different from 25 °C with p < 0.01.
Figure 4
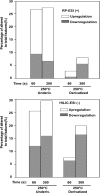
Percentage of altered (both upregulated and downregulated) features to the total features in the underivatized and derivatized 64 small molecule mixture heated with 250 °C for 30, 60, and 300 s using both the RP-ESI (+) and HILIC-ESI (−) methods.
Figure 5
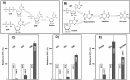
Proposed thermal degradation pathways for: (A) UTP, UDP, UMP, and uridine and (B) IMP, inosine, hypoxanthine, xanthine, and uric acid. Additionally, the relative increases of: (C) UTP, UDP, and UMP; D) ATP, ADP, and AMP; and (E) inosine, hypoxanthine (HPX), and uric acid heated at 250 °C for 60 s compared with 25 °C are shown. Error bars show standard deviation (based on triplicates).
Similar articles
- Thermal degradation of metabolites in urine using multiple isotope-labelled internal standards for off-line GC metabolomics - effects of injector and oven temperatures.
Tsochatzis ED, Nebel C, Danielsen M, Sundekilde UK, Kastrup Dalsgaard T. Tsochatzis ED, et al. J Chromatogr B Analyt Technol Biomed Life Sci. 2021 Sep 1;1181:122902. doi: 10.1016/j.jchromb.2021.122902. Epub 2021 Sep 4. J Chromatogr B Analyt Technol Biomed Life Sci. 2021. PMID: 34530307 - Investigation of the derivatization conditions for GC-MS metabolomics of biological samples.
Moros G, Chatziioannou AC, Gika HG, Raikos N, Theodoridis G. Moros G, et al. Bioanalysis. 2017 Jan;9(1):53-65. doi: 10.4155/bio-2016-0224. Bioanalysis. 2017. PMID: 27921459 - Simultaneous quantification of purine and pyrimidine bases, nucleosides and their degradation products in bovine blood plasma by high performance liquid chromatography tandem mass spectrometry.
Stentoft C, Vestergaard M, Løvendahl P, Kristensen NB, Moorby JM, Jensen SK. Stentoft C, et al. J Chromatogr A. 2014 Aug 22;1356:197-210. doi: 10.1016/j.chroma.2014.06.065. Epub 2014 Jun 27. J Chromatogr A. 2014. PMID: 25017393 - Stable isotope coded derivatizing reagents as internal standards in metabolite profiling.
Bruheim P, Kvitvang HF, Villas-Boas SG. Bruheim P, et al. J Chromatogr A. 2013 Jun 28;1296:196-203. doi: 10.1016/j.chroma.2013.03.072. Epub 2013 Apr 9. J Chromatogr A. 2013. PMID: 23628173 Review. - Oxidized and nitrated oleic acid in biological systems: analysis by GC-MS/MS and LC-MS/MS, and biological significance.
Tsikas D, Zoerner AA, Jordan J. Tsikas D, et al. Biochim Biophys Acta. 2011 Nov;1811(11):694-705. doi: 10.1016/j.bbalip.2011.06.015. Epub 2011 Jun 26. Biochim Biophys Acta. 2011. PMID: 21771665 Review.
Cited by
- Uterine lumen fluid is metabolically semi-autonomous.
Simintiras CA, Drum JN, Liu H, Sofia Ortega M, Spencer TE. Simintiras CA, et al. Commun Biol. 2022 Mar 1;5(1):191. doi: 10.1038/s42003-022-03134-0. Commun Biol. 2022. PMID: 35233029 Free PMC article. - A Review of the Botany, Traditional Use, Phytochemistry, Analytical Methods, Pharmacological Effects, and Toxicity of Angelicae Pubescentis Radix.
Yang L, Hou A, Wang S, Zhang J, Man W, Guo X, Yang B, Jiang H, Kuang H, Wang Q. Yang L, et al. Evid Based Complement Alternat Med. 2020 Jul 31;2020:7460781. doi: 10.1155/2020/7460781. eCollection 2020. Evid Based Complement Alternat Med. 2020. PMID: 32831877 Free PMC article. Review. - JPA: Joint Metabolic Feature Extraction Increases the Depth of Chemical Coverage for LC-MS-Based Metabolomics and Exposomics.
Guo J, Shen S, Liu M, Wang C, Low B, Chen Y, Hu Y, Xing S, Yu H, Gao Y, Fang M, Huan T. Guo J, et al. Metabolites. 2022 Feb 26;12(3):212. doi: 10.3390/metabo12030212. Metabolites. 2022. PMID: 35323655 Free PMC article. - Preliminary Investigation of the Effect of Maceration Procedures on Bone Metabolome and Lipidome.
Bonicelli A, Cheung W, Hughes S, Wescott DJ, Procopio N. Bonicelli A, et al. Metabolites. 2022 Oct 25;12(11):1020. doi: 10.3390/metabo12111020. Metabolites. 2022. PMID: 36355103 Free PMC article. - Decoding the metabolic response of Escherichia coli for sensing trace heavy metals in water.
Wei H, Huang Y, Santiago PJ, Labachyan KE, Ronaghi S, Banda Magana MP, Huang YH, C Jiang S, Hochbaum AI, Ragan R. Wei H, et al. Proc Natl Acad Sci U S A. 2023 Feb 14;120(7):e2210061120. doi: 10.1073/pnas.2210061120. Epub 2023 Feb 6. Proc Natl Acad Sci U S A. 2023. PMID: 36745806 Free PMC article.
References
- Bundy J. G.; Davey M. P.; Viant M. R. Metabolomics 2009, 5, 3–2110.1007/s11306-008-0152-0. - DOI
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous