Algal ancestor of land plants was preadapted for symbiosis - PubMed (original) (raw)
. 2015 Oct 27;112(43):13390-5.
doi: 10.1073/pnas.1515426112. Epub 2015 Oct 5.
Guru V Radhakrishnan 2, Dhileepkumar Jayaraman 3, Jitender Cheema 2, Mathilde Malbreil 4, Jeremy D Volkening 5, Hiroyuki Sekimoto 6, Tomoaki Nishiyama 7, Michael Melkonian 8, Lisa Pokorny 9, Carl J Rothfels 10, Heike Winter Sederoff 11, Dennis W Stevenson 12, Barbara Surek 8, Yong Zhang 13, Michael R Sussman 5, Christophe Dunand 4, Richard J Morris 2, Christophe Roux 4, Gane Ka-Shu Wong 14, Giles E D Oldroyd 2, Jean-Michel Ané 3
Affiliations
- PMID: 26438870
- PMCID: PMC4629359
- DOI: 10.1073/pnas.1515426112
Algal ancestor of land plants was preadapted for symbiosis
Pierre-Marc Delaux et al. Proc Natl Acad Sci U S A. 2015.
Abstract
Colonization of land by plants was a major transition on Earth, but the developmental and genetic innovations required for this transition remain unknown. Physiological studies and the fossil record strongly suggest that the ability of the first land plants to form symbiotic associations with beneficial fungi was one of these critical innovations. In angiosperms, genes required for the perception and transduction of diffusible fungal signals for root colonization and for nutrient exchange have been characterized. However, the origin of these genes and their potential correlation with land colonization remain elusive. A comprehensive phylogenetic analysis of 259 transcriptomes and 10 green algal and basal land plant genomes, coupled with the characterization of the evolutionary path leading to the appearance of a key regulator, a calcium- and calmodulin-dependent protein kinase, showed that the symbiotic signaling pathway predated the first land plants. In contrast, downstream genes required for root colonization and their specific expression pattern probably appeared subsequent to the colonization of land. We conclude that the most recent common ancestor of extant land plants and green algae was preadapted for symbiotic associations. Subsequent improvement of this precursor stage in early land plants through rounds of gene duplication led to the acquisition of additional pathways and the ability to form a fully functional arbuscular mycorrhizal symbiosis.
Keywords: algae; phylogeny; plant evolution; plant–microbe interactions; symbiosis.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
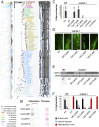
Fig. 1.
The green lineage and the AM symbiosis. (A) Schematic representation of the green lineage. Light blue: chlorophytes and basal charophytes; dark blue: advanced charophytes. Green bars on the left indicate the actual number of transcriptomes and genomes included in the extended dataset used in this study. (B) Schematic representation of the symbiotic genes.
Fig. 2.
Stepwise evolution of CCaMK in the green lineage. (A) Maximum-likelihood phylogenetic tree of CCaMK extracted from the CDPK tree on the left. A high-resolution image of the CDPK tree is available in SI Appendix, Fig. S5. Colored boxes on the right indicate the conservation of the CBD within the CCaMK clade compared with MtCCaMK. Black, blue, and white squares indicate identical, homologous, and different amino acids, respectively. (B) Calmodulin-binding activity of MtCCaMK, CpCCaMK, and two chlorophyte CDPKs (CrCDPK1 and CvCDPK3) in the presence or absence of calcium. This assay is positive for MtCCaMK and CpCCaMK (dark spot) and negative for CrCDPK1 and CvCDPK3. The original plots are available in SI Appendix, Fig. S20. (C) Transcomplementation assays of the Mtccamk-1 TRV25 mutant with various CCaMKs and CDPKs including NpCCaMK and CaCCaMK. Letters indicate statistically supported groups according to ANOVA and Tukey’s honestly significant difference tests (P < 0.01). EV, empty vector. (D) Representative pictures of wheat germ agglutinin-FITC–stained AM fungi colonizing plant roots in the transcomplementation assay. (Scale bars, 100 μm.) (E) Amplification by RT-PCR of the symbiotic phosphate transporter MtPT4 and ubiquitin (MtUBI). (F) Quantification of the hyphopodia leading to internal hyphae and arbuscules.
Fig. 3.
Multiple paths for the evolution of symbiotic genes in basal land plants. (A) Maximum-likelihood phylogenetic tree of NSP2, RAM1, RAD1, and NSP1 extracted from the GRAS tree on the left. These four clades originate from land plant-specific duplications. (B) Maximum-likelihood phylogenetic tree of HA1 and liverworts HA extracted from the H+-ATPase tree (available in SI Appendix, Fig. S17). (C) Differential expression of LcNSP2, LcRAM1, LcRAD1, LcNSP1, and LcHA in mock-treated (Mock) or colonized (AM) Lunularia cruciata. *P < 0.05. AM-inducible H +- ATPase evolved independently in angiosperms and liverworts.
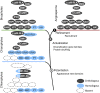
Fig. 4.
Stepwise acquisition of symbiotic genes in the green lineage. The appearance of new domains in advanced charophytes led to the emergence of new protein families and to the evolution of the symbiotic signaling module via the neofunctionalization of existing proteins (DMI1 and CCaMK). Most of the downstream symbiotic genes evolved in basal land plants via gene duplication in these newly evolved gene families (i.e., STR) or by combining existing domains in new proteins (i.e., VAPYRIN). Finally, genes originating from lineage-specific duplications were recruited independently to fulfill their symbiotic functions (PT and H+-ATPase).
Similar articles
- Evolutionary History of Subtilases in Land Plants and Their Involvement in Symbiotic Interactions.
Taylor A, Qiu YL. Taylor A, et al. Mol Plant Microbe Interact. 2017 Jun;30(6):489-501. doi: 10.1094/MPMI-10-16-0218-R. Epub 2017 May 8. Mol Plant Microbe Interact. 2017. PMID: 28353400 - Evolution of endosymbiosis-mediated nuclear calcium signaling in land plants.
Lam AHC, Cooke A, Wright H, Lawson DM, Charpentier M. Lam AHC, et al. Curr Biol. 2024 May 20;34(10):2212-2220.e7. doi: 10.1016/j.cub.2024.03.063. Epub 2024 Apr 19. Curr Biol. 2024. PMID: 38642549 - The microRNA miR171h modulates arbuscular mycorrhizal colonization of Medicago truncatula by targeting NSP2.
Lauressergues D, Delaux PM, Formey D, Lelandais-Brière C, Fort S, Cottaz S, Bécard G, Niebel A, Roux C, Combier JP. Lauressergues D, et al. Plant J. 2012 Nov;72(3):512-22. doi: 10.1111/j.1365-313X.2012.05099.x. Epub 2012 Aug 30. Plant J. 2012. PMID: 22775306 - Major transitions in the evolution of early land plants: a bryological perspective.
Ligrone R, Duckett JG, Renzaglia KS. Ligrone R, et al. Ann Bot. 2012 Apr;109(5):851-71. doi: 10.1093/aob/mcs017. Epub 2012 Feb 22. Ann Bot. 2012. PMID: 22356739 Free PMC article. Review. - Symbiotic options for the conquest of land.
Field KJ, Pressel S, Duckett JG, Rimington WR, Bidartondo MI. Field KJ, et al. Trends Ecol Evol. 2015 Aug;30(8):477-86. doi: 10.1016/j.tree.2015.05.007. Epub 2015 Jun 22. Trends Ecol Evol. 2015. PMID: 26111583 Review.
Cited by
- DWARF27 and CAROTENOID CLEAVAGE DIOXYGENASE 7 genes regulate release, germination and growth of gemma in Marchantia polymorpha.
Jibran R, Tahir J, Andre CM, Janssen BJ, Drummond RSM, Albert NW, Zhou Y, Davies KM, Snowden KC. Jibran R, et al. Front Plant Sci. 2024 Jun 25;15:1358745. doi: 10.3389/fpls.2024.1358745. eCollection 2024. Front Plant Sci. 2024. PMID: 38984156 Free PMC article. - Evolution of the Symbiosis-Specific GRAS Regulatory Network in Bryophytes.
Grosche C, Genau AC, Rensing SA. Grosche C, et al. Front Plant Sci. 2018 Nov 6;9:1621. doi: 10.3389/fpls.2018.01621. eCollection 2018. Front Plant Sci. 2018. PMID: 30459800 Free PMC article. - Neighboring plants divergently modulate effects of loss-of-function in maize mycorrhizal phosphate uptake on host physiology and root fungal microbiota.
Fabiańska I, Pesch L, Koebke E, Gerlach N, Bucher M. Fabiańska I, et al. PLoS One. 2020 Jun 17;15(6):e0232633. doi: 10.1371/journal.pone.0232633. eCollection 2020. PLoS One. 2020. PMID: 32555651 Free PMC article. - The receptor-like kinase ARK controls symbiotic balance across land plants.
Sgroi M, Hoey D, Medina Jimenez K, Bowden SL, Hope M, Wallington EJ, Schornack S, Bravo A, Paszkowski U. Sgroi M, et al. Proc Natl Acad Sci U S A. 2024 Jul 23;121(30):e2318982121. doi: 10.1073/pnas.2318982121. Epub 2024 Jul 16. Proc Natl Acad Sci U S A. 2024. PMID: 39012828 Free PMC article. - Biotic interactions, evolutionary forces and the pan-plant specialized metabolism.
de Vries S, Feussner I. de Vries S, et al. Philos Trans R Soc Lond B Biol Sci. 2024 Nov 18;379(1914):20230362. doi: 10.1098/rstb.2023.0362. Epub 2024 Sep 30. Philos Trans R Soc Lond B Biol Sci. 2024. PMID: 39343027 Free PMC article. Review.
References
- Beerling D. The Emerald Planet. How Plants Changed Earth’s History. Oxford Univ Press; Oxford: 2007.
- Lal R. Soil carbon sequestration impacts on global climate change and food security. Science. 2004;304(5677):1623–1627. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- BB/L014130/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BBS/E/J/00000611/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials