ATM Dependent Silencing Links Nucleolar Chromatin Reorganization to DNA Damage Recognition - PubMed (original) (raw)
ATM Dependent Silencing Links Nucleolar Chromatin Reorganization to DNA Damage Recognition
Shane M Harding et al. Cell Rep. 2015.
Abstract
Resolution of DNA double-strand breaks (DSBs) is essential for the suppression of genome instability. DSB repair in transcriptionally active genomic regions represents a unique challenge that is associated with ataxia telangiectasia mutated (ATM) kinase-mediated transcriptional silencing. Despite emerging insights into the underlying mechanisms, how DSB silencing connects to DNA repair remains undefined. We observe that silencing within the rDNA depends on persistent DSBs. Non-homologous end-joining was the predominant mode of DSB repair allowing transcription to resume. ATM-dependent rDNA silencing in the presence of persistent DSBs led to the large-scale reorganization of nucleolar architecture, with movement of damaged chromatin to nucleolar cap regions. These findings identify ATM-dependent temporal and spatial control of DNA repair and provide insights into how communication between DSB signaling and ongoing transcription promotes genome integrity.
Copyright © 2015 The Authors. Published by Elsevier Inc. All rights reserved.
Figures
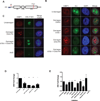
Figure 1. Inhibition of NHEJ exacerbates ATM-dependent silencing of rDNA transcription
(A) Schematic drawing of a nascent 45S rRNA transcript. The 45S transcript is rapidly cleaved at A0 as indicated by the red arrow. The blue bar indicates the amplicon monitored by RT-qPCR. The I-PpoI cut site within the 28S region is indicated. (B) Immunofluorescence of 53BP1 (red) and nascent transcription measured by 5-Ethynyl uridine (EU) incorporation following I-PpoI induction in the presence of the indicated inhibitors. A merge overlaid with DAPI is shown. Scale bar is 10µm. (C) Combined immunofluorescence of 53BP1 (red) and nascent rRNA measured by RNA-FISH. Scale bar is 10µm. (D) MCF10A cells were treated with the indicated siRNA for 72h before I-PpoI induction and nascent transcription was monitored 5 hr later. Error bars represent SEM. *p<0.05. **p<0.01, ***p<0.001 (E) RT-qPCR measurement of nascent 45S transcript following induction of I-PpoI endonuclease for 5h in the presence of the indicated inhibitors in MCF10A cells. Please see supplemental experimental procedures for inhibitor details. Error bars represent SEM of at least 3 biological replicates. ***p<0.001
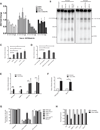
Figure 2. Persistent DSBs are required for ATM-dependent rDNA silencing
(A) Nascent 45S transcription was measured as in Figure 1B at the indicated times following I-PpoI induction. Error bars represent SEM of 3 biological replicates (B) PFGE and hybridization of rDNA probes following I-PpoI induction with indicated treatments. Approximate sizes are indicated based on DNA markers. (C) Quantification of PFGE hybridization signal for inhibitor treated MCF10A cells as indicated. (D) Quantification of PFGE hybridization signal for siRNA treated MCF10A cells as indicated. (E) Quantification of PFGE hybridization signal for MEFs either untreated or transfected with I-PpoI mRNA. (F) Quantification as in (E), except CreERT2-expressing wild-type and conditionally deleted Brca1f/f were assayed. (G) Fraction of EU-positive cells before and 20h following I-PpoI mRNA transfection. ***p<0.001. (H) Cellular viability was measured by the MTS assay at 40h following washout of transfected I-PpoI mRNA. All error bars represent SEM.
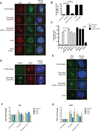
Figure 3. ATM-dependent DSB silencing leads to nucleolar reorganization
(A) Immunofluorescence of UBF (red) and nascent transcription as measured in Figure 1D. Actinomycin D treatment serves as a control for cap formation in the absence of I-PpoI induction. Scale bar is 10µm. (B) Quantification of the percentage of cells with UBF nucleolar caps for the experiment described in (A). Error bar represents SEM of at least 3 biological replicates. ***p<0.001 (C) Percentage of cells with UBF nucleolar caps following I-PpoI induction 72h after siRNA transfection. *p<0.05, ***p<0.001 (D) Immunofluorescent staining of 53BP1 (green) and Nucleophosmin (NPM). Scale bar is 10µm. (E) DNA-FISH of rDNA following I-PpoI induction. Scale bar is 10µm. (F) Fold change of ChIP-qPCR signal for UBF as indicated following induction of I-PpoI in the presence of indicated inhibitors. Primer sets were targeted to the promoter, 28S coding region or the intergenic spacer (IGS) between rDNA repeats. Error bars represent SEM of 3 biological replicates. (G) Fold change of ChIP-qPCR signal for 53BP1 as in Figure 3F. Error bars represent SEM of 3 biological replicates.
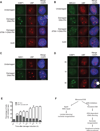
Figure 4. Nucleolar DSBs are recognized following ATM-dependent nucleolar cap formation
(A) Immunofluorescence staining of 53BP1 (green) and UBF (red). Merged images were overlaid with DAPI-stained nuclei. Inset depicts a 2× digital zoom of 53BP1 juxtaposition to UBF at nucleolar caps. Representative images of 3 biological replicates are shown. Scale bar is 10µm. (B) Immunofluorecence of BRCA1 (green) and UBF (red) as in Figure 4A. (C) Immunofluorecence of NBS1 (green) and UBF (red) as in Figure 4A. (D) Immunofluorescence of 53BP1 (green) and UBF (red) at 0hr, 1 hr and 5 hr following I-PpoI induction in the presence of DNA-PK inhibitor. An arrowhead indicates coalesced UBF. An asterisk indicates 53BP1-associated UBF nucleolar caps. Scale bar is 10µm. (E) Quantification of Figure 4E. Cells displaying punctate non-53BP1-associated UBF (Arrowhead in Figure 4D) and 53BP1-associated UBF nucleolar caps (Asterisk in Figure 4D) were quantified at indicated times following I-PpoI induction in the presence of DNA-PK inhibitor. Error bars represent SEM for 3 biological replicates. (F) Schematic of rDNA DSB silencing. As described in the text, persistent DSBs exact ATM dependent nucleolar reorganization and DSB recognition subsequent to transcriptional silencing of rDNA loci.
Similar articles
- Double-strand breaks in ribosomal RNA genes activate a distinct signaling and chromatin response to facilitate nucleolar restructuring and repair.
Korsholm LM, Gál Z, Lin L, Quevedo O, Ahmad DA, Dulina E, Luo Y, Bartek J, Larsen DH. Korsholm LM, et al. Nucleic Acids Res. 2019 Sep 5;47(15):8019-8035. doi: 10.1093/nar/gkz518. Nucleic Acids Res. 2019. PMID: 31184714 Free PMC article. - Nucleolar responses to DNA double-strand breaks.
Larsen DH, Stucki M. Larsen DH, et al. Nucleic Acids Res. 2016 Jan 29;44(2):538-44. doi: 10.1093/nar/gkv1312. Epub 2015 Nov 28. Nucleic Acids Res. 2016. PMID: 26615196 Free PMC article. Review. - The heterochromatic barrier to DNA double strand break repair: how to get the entry visa.
Goodarzi AA, Jeggo PA. Goodarzi AA, et al. Int J Mol Sci. 2012;13(9):11844-11860. doi: 10.3390/ijms130911844. Epub 2012 Sep 19. Int J Mol Sci. 2012. PMID: 23109886 Free PMC article. Review. - A DYRK1B-dependent pathway suppresses rDNA transcription in response to DNA damage.
Dong C, An L, Yu CH, Huen MSY. Dong C, et al. Nucleic Acids Res. 2021 Feb 22;49(3):1485-1496. doi: 10.1093/nar/gkaa1290. Nucleic Acids Res. 2021. PMID: 33469661 Free PMC article. - Phenotypic Analysis of ATM Protein Kinase in DNA Double-Strand Break Formation and Repair.
Mian E, Wiesmüller L. Mian E, et al. Methods Mol Biol. 2017;1599:317-334. doi: 10.1007/978-1-4939-6955-5_23. Methods Mol Biol. 2017. PMID: 28477129
Cited by
- Nucleolar stress: Friend or foe in cardiac function?
Yan D, Hua L. Yan D, et al. Front Cardiovasc Med. 2022 Oct 31;9:1045455. doi: 10.3389/fcvm.2022.1045455. eCollection 2022. Front Cardiovasc Med. 2022. PMID: 36386352 Free PMC article. Review. - The Secret Life of Chromosome Loops upon DNA Double-Strand Break.
Arnould C, Legube G. Arnould C, et al. J Mol Biol. 2020 Feb 7;432(3):724-736. doi: 10.1016/j.jmb.2019.07.036. Epub 2019 Aug 8. J Mol Biol. 2020. PMID: 31401119 Free PMC article. Review. - Arp2/3 and Unc45 maintain heterochromatin stability in Drosophila polytene chromosomes.
Dialynas G, Delabaere L, Chiolo I. Dialynas G, et al. Exp Biol Med (Maywood). 2019 Nov;244(15):1362-1371. doi: 10.1177/1535370219862282. Epub 2019 Jul 31. Exp Biol Med (Maywood). 2019. PMID: 31364400 Free PMC article. - The chemotherapeutic agent CX-5461 irreversibly blocks RNA polymerase I initiation and promoter release to cause nucleolar disruption, DNA damage and cell inviability.
Mars JC, Tremblay MG, Valere M, Sibai DS, Sabourin-Felix M, Lessard F, Moss T. Mars JC, et al. NAR Cancer. 2020 Dec;2(4):zcaa032. doi: 10.1093/narcan/zcaa032. Epub 2020 Nov 6. NAR Cancer. 2020. PMID: 33196044 Free PMC article. - Treacle Sticks the Nucleolar Responses to DNA Damage Together.
Gál Z, Nieto B, Boukoura S, Rasmussen AV, Larsen DH. Gál Z, et al. Front Cell Dev Biol. 2022 May 12;10:892006. doi: 10.3389/fcell.2022.892006. eCollection 2022. Front Cell Dev Biol. 2022. PMID: 35646927 Free PMC article. Review.
References
- Berkovich E, Monnat RJ, Kastan MB. Roles of ATM and NBS1 in chromatin structure modulation and DNA double-strand break repair. Nat. Cell Biol. 2007;9:683–690. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- CA138835/CA/NCI NIH HHS/United States
- R01 CA174904/CA/NCI NIH HHS/United States
- R01 GM101149/GM/NIGMS NIH HHS/United States
- CA17494/CA/NCI NIH HHS/United States
- R01 CA138835/CA/NCI NIH HHS/United States
- GM101149/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous