Structure of Ljungan virus provides insight into genome packaging of this picornavirus - PubMed (original) (raw)
Xiangxi Wang 3, Jingshan Ren 1, Claudine Porta 1 2, Hannah Wenham 2, Jens-Ola Ekström 4, Anusha Panjwani 2, Nick J Knowles 2, Abhay Kotecha 1, C Alistair Siebert 1, A Michael Lindberg 4, Elizabeth E Fry 1, Zihe Rao 3 5, Tobias J Tuthill 2, David I Stuart 1 6
Affiliations
- PMID: 26446437
- PMCID: PMC4633645
- DOI: 10.1038/ncomms9316
Structure of Ljungan virus provides insight into genome packaging of this picornavirus
Ling Zhu et al. Nat Commun. 2015.
Abstract
Picornaviruses are responsible for a range of human and animal diseases, but how their RNA genome is packaged remains poorly understood. A particularly poorly studied group within this family are those that lack the internal coat protein, VP4. Here we report the atomic structure of one such virus, Ljungan virus, the type member of the genus Parechovirus B, which has been linked to diabetes and myocarditis in humans. The 3.78-Å resolution cryo-electron microscopy structure shows remarkable features, including an extended VP1 C terminus, forming a major protuberance on the outer surface of the virus, and a basic motif at the N terminus of VP3, binding to which orders some 12% of the viral genome. This apparently charge-driven RNA attachment suggests that this branch of the picornaviruses uses a different mechanism of genome encapsidation, perhaps explored early in the evolution of picornaviruses.
Figures
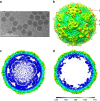
Figure 1. Single-particle 3D reconstructions of LV.
(a) Cryo-EM image of LV. (b) 3D reconstruction of LV particle viewed down the three-fold axis. The surface is coloured by radius from blue through green to red from the lowest to the highest radius. The icosahedral five-fold, three-fold and two-fold axes are labelled as 5, 3, and 2, respectively. (c) A thin slice of the central section of the LV particle viewed down the three-fold axis. (d) The central section of the LV reconstruction rendered at higher contour level (4σ above the mean), viewed along a five-fold axis of symmetry and depth queued to reveal the five high-density protrusions inside the capsid under each vertex.
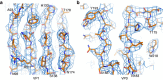
Figure 2. Electron density maps.
(a) From a section of VP1. (b) From a section of VP0.
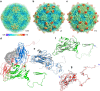
Figure 3. Overall structure of LV.
(a–c) Comparison of capsids from LV (b) with HAV (a) and TrV (c). The capsids are coloured by radius from blue to red according to the scale bar shown. (d) Ribbon diagram of the LV protomer. VP1, VP0 and VP3 are shown in blue, green and red, respectively; the VP1 C-terminal 55 residues that form the surface protrusion around the five-fold axis are shown by a lower-resolution electron density map. (e–g) Ribbon diagrams showing the LV capsid proteins VP1 (e), VP0 (f) and VP3 (g) with the individual strands and termini labelled, compared with those of HAV (grey), the N- and C termini of the LV proteins are labelled in blue and red, respectively.
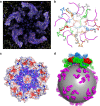
Figure 4. The partial structure of the LV genome and virus encapsidation.
(a) Electron density for the N termini of VP3 and the ordered RNA around the five-fold axis. (b) The structure of VP3 N termini around the five-fold axis (individual copies distinguished as blue, green, red, orange and grey ribbons, from residue 4 labelled N to residue 64 labelled C) and their proximity to the RNA (magenta). (c) Inner electrostatic surface of the LV pentamer showing the predominance of positive charge that interacts with the backbone of the genomic RNA (magenta sticks). (d) Cartoon diagram representing the LV genomic RNA as a sphere with ordered fragments forming pentameric protrusions (magenta); the pentamers of capsid protein associate with the genome RNA through electrostatic interaction.
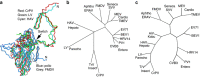
Figure 5. Phylogeny.
(a) Superposition of LV VP0 with VP2 of poliovirus, FMDV, HAV and CrPV. The N terminus of LV VP0 folds similarly to that of HAV and CrPV but differently to other picornaviruses. The star marks the EF loop that forms part of the south wall of the canyon in poliovirus. (b) Structure and (c) sequence-based phylogenetic trees of representative picornaviruses and cripaviruses, respectively: CVB3, coxsackievirus B3; PV1, poliovirus type 1; HRV14, human rhinovirus 14; BEV1, bovine enterovirus type 1; EV71, EV-A71; TMEV, Theilers virus; MEV, Mengo virus; SVV, Seneca valley virus; FMDV, foot-and-mouth disease virus; ERAV, equine rhinitis A virus; HAV, hepatitis A virus; AiV, Aichi virus; TrV, triatoma virus; CrPV, cricket paralysis virus.
Similar articles
- Genome characterisation of two Ljungan virus isolates from wild bank voles (Myodes glareolus) in Sweden.
Pounder KC, Watts PC, Niklasson B, Kallio ERK, Marston DA, Fooks AR, Begon M, McElhinney LM. Pounder KC, et al. Infect Genet Evol. 2015 Dec;36:156-164. doi: 10.1016/j.meegid.2015.09.010. Epub 2015 Sep 13. Infect Genet Evol. 2015. PMID: 26375731 - The Structure of Human Parechovirus 1 Reveals an Association of the RNA Genome with the Capsid.
Kalynych S, Pálková L, Plevka P. Kalynych S, et al. J Virol. 2015 Nov 18;90(3):1377-86. doi: 10.1128/JVI.02346-15. Print 2016 Feb 1. J Virol. 2015. PMID: 26581987 Free PMC article. - Mechanisms of assembly and genome packaging in an RNA virus revealed by high-resolution cryo-EM.
Hesketh EL, Meshcheriakova Y, Dent KC, Saxena P, Thompson RF, Cockburn JJ, Lomonossoff GP, Ranson NA. Hesketh EL, et al. Nat Commun. 2015 Dec 10;6:10113. doi: 10.1038/ncomms10113. Nat Commun. 2015. PMID: 26657148 Free PMC article. - Human parechoviruses--biology and clinical significance.
Stanway G, Joki-Korpela P, Hyypiä T. Stanway G, et al. Rev Med Virol. 2000 Jan-Feb;10(1):57-69. doi: 10.1002/(sici)1099-1654(200001/02)10:1<57::aid-rmv266>3.0.co;2-h. Rev Med Virol. 2000. PMID: 10654005 Review. - Properties and classification of hepatitis A virus.
Melnick JL. Melnick JL. Vaccine. 1992;10 Suppl 1:S24-6. doi: 10.1016/0264-410x(92)90536-s. Vaccine. 1992. PMID: 1335653 Review.
Cited by
- Structure of human Aichi virus and implications for receptor binding.
Zhu L, Wang X, Ren J, Kotecha A, Walter TS, Yuan S, Yamashita T, Tuthill TJ, Fry EE, Rao Z, Stuart DI. Zhu L, et al. Nat Microbiol. 2016 Sep 5;1(11):16150. doi: 10.1038/nmicrobiol.2016.150. Nat Microbiol. 2016. PMID: 27595320 - Structures of Coxsackievirus A10 unveil the molecular mechanisms of receptor binding and viral uncoating.
Zhu L, Sun Y, Fan J, Zhu B, Cao L, Gao Q, Zhang Y, Liu H, Rao Z, Wang X. Zhu L, et al. Nat Commun. 2018 Nov 26;9(1):4985. doi: 10.1038/s41467-018-07531-0. Nat Commun. 2018. PMID: 30478256 Free PMC article. - Capsid Structure of a Marine Algal Virus of the Order Picornavirales.
Munke A, Kimura K, Tomaru Y, Okamoto K. Munke A, et al. J Virol. 2020 Apr 16;94(9):e01855-19. doi: 10.1128/JVI.01855-19. Print 2020 Apr 16. J Virol. 2020. PMID: 32024776 Free PMC article. - A comparative analysis of parechovirus protein structures with other picornaviruses.
Domanska A, Guryanov S, Butcher SJ. Domanska A, et al. Open Biol. 2021 Jul;11(7):210008. doi: 10.1098/rsob.210008. Epub 2021 Jul 28. Open Biol. 2021. PMID: 34315275 Free PMC article. - An outbreak of severe infections among Australian infants caused by a novel recombinant strain of human parechovirus type 3.
Nelson TM, Vuillermin P, Hodge J, Druce J, Williams DT, Jasrotia R, Alexandersen S. Nelson TM, et al. Sci Rep. 2017 Mar 14;7:44423. doi: 10.1038/srep44423. Sci Rep. 2017. PMID: 28290509 Free PMC article.
References
- Rao A. L. Genome packaging by spherical plant RNA viruses. Annu. Rev. Phytopathol. 44, 61–87 (2006). - PubMed
- Casjens S. R. The DNA-packaging nanomotor of tailed bacteriophages. Nat. Rev. Microbiol. 9, 647–657 (2011). - PubMed
- Whitton J. L., Cornell C. T. & Feuer R. Host and virus determinants of picornavirus pathogenesis and tropism. Nat. Rev. Microbiol. 3, 765–776 (2005). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- G100099/MRC_/Medical Research Council/United Kingdom
- 093305/Z/10/Z/WT_/Wellcome Trust/United Kingdom
- MR/K020811/1/MRC_/Medical Research Council/United Kingdom
- BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- MR/N00065X/1/MRC_/Medical Research Council/United Kingdom
- G1100525/MRC_/Medical Research Council/United Kingdom
- 090532/WT_/Wellcome Trust/United Kingdom
- G1000099/MRC_/Medical Research Council/United Kingdom
- 060208/Z/00/Z/WT_/Wellcome Trust/United Kingdom
- 09053/Z/09/Z/WT_/Wellcome Trust/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources