MicroRNA 140 Promotes Expression of Long Noncoding RNA NEAT1 in Adipogenesis - PubMed (original) (raw)
MicroRNA 140 Promotes Expression of Long Noncoding RNA NEAT1 in Adipogenesis
Ramkishore Gernapudi et al. Mol Cell Biol. 2015.
Abstract
More than 40% of the U.S. population are clinically obese and suffer from metabolic syndrome with an increased risk of postmenopausal estrogen receptor-positive breast cancer. Adipocytes are the primary component of adipose tissue and are formed through adipogenesis from precursor mesenchymal stem cells. While the major molecular pathways of adipogenesis are understood, little is known about the noncoding RNA signaling networks involved in adipogenesis. Using adipocyte-derived stem cells (ADSCs) isolated from wild-type and microRNA 140 (miR-140) knockout mice, we identify a novel miR-140/long noncoding RNA (lncRNA) NEAT1 signaling network necessary for adipogenesis. miR-140 knockout ADSCs have dramatically decreased adipogenic capabilities associated with downregulation of NEAT1 expression. We identified a miR-140 binding site in NEAT1 and found that mature miR-140 in the nucleus can physically interact with NEAT1, leading to increased NEAT1 expression. We demonstrated that reexpression of NEAT1 in miR-140 knockout ADSCs is sufficient to restore their ability to undergo differentiation. Our results reveal an exciting new noncoding RNA signaling network that regulates adipogenesis and that is a potential new target in the prevention or treatment of obesity.
Copyright © 2015, American Society for Microbiology. All Rights Reserved.
Figures
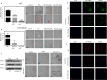
FIG 1
SFN treatment inhibits preadipocyte differentiation in primary SVF. (A) Sulforaphane treatment (5 or 10 μM) resulted in a dramatic decrease in differentiation, as evidenced by a decrease in lipid droplet accumulation. Primary SVF following differentiation (the 9th day after stimulation with
d
-biotin, dexamethasone, insulin, and IBMX cocktail) was stained with Oil Red O. (B) Primary SVF obtained from miR-140 KO mice showed a decrease in differentiation in both the presence and absence of SFN (5 or 10 μM). Oil Red O staining indicated that the knockout of miR-140 resulted in a decrease in differentiation, and SFN treatment (5 or 10 μM) blocked adipocyte differentiation. (C) Protein analysis of differentiated WT and miR-140 KO adipocytes for adipocyte markers CEBP/α and PPARγ and IHC imaging of mammary tissue sections from WT and miR-140 KO mice for adipocyte markers, CEBP/α, PPARγ, and adiponectin. Brown precipitate was developed using an avidin-biotin peroxidase substrate kit (Vector Laboratories, CA). Nuclei were counterstained with hematoxylin. The images were captured using a Nikon-Ti microscope. (D and E) Immunofluorescence imaging of differentiated adipocytes. The SVF from miR-140 KO was allowed to differentiate in an adipogenic cocktail with or without SFN (5 or 10 μM) for 10 days. After differentiation, cells were fixed in 4% paraformaldehyde (PFA) and analyzed for the expression levels of mature adipocyte markers (CEBP/α and PPARγ). The cells were stained with anti-rabbit Alexa 488- or 555-conjugated secondary antibody, and nuclei were counterstained with DAPI. Shown are representative images from experiments done in triplicate. The scale bars represent 100 μm. The data represent means and SE.
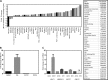
FIG 2
Differential expression of lncRNAs in adipocytes compared to preadipocytes. (A) A 96-well-based human Lnc profiler qPCR array consisting of 90 different lncRNAs, along with 5 housekeeping genes and one negative control, was used to screen adipocytes. The results for the genes that are either upregulated or downregulated by at least 1.5-fold are listed in the table. (B) Expression levels of NEAT1 in preadipocytes (ADSCs) and differentiated adipocytes (DA) compared to ATCC (human lung fibroblasts) and DCIS (MCF10DCIS). (C) Comparative analysis of lncRNA identified by NCBI reference number (NR). The data represent means and SE (n = 3).
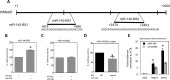
FIG 3
The miR-140 binding sites within mouse NEAT1 (mNeat1) were identified using RNAhybrid 2.2. (A) Target binding sites of miR-140 (BS1 and BS2) within NEAT1. (B) Luciferase reporter assays were performed and confirmed that ectopic expression of miR-140 increased the activity of the BS1 reporter. (C) Ectopic expression of miR-140 has no impact on BS2. (D) Mutation of BS1 inhibited miR-140 induction of luciferase activity. (E) Biotin-labeled miR-140 pulldown demonstrated binding of miR-140 to NEAT1. ACTB, β-actin. The data represent means and SE (n = 3). *, P < 0.05.
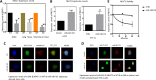
FIG 4
Expression levels of NEAT1 by RT-PCR analysis and validation of miR-140 downregulation in preadipocytes. (A) Comparison of NEAT1 expression levels in WT and miR-140 KO adipocyte-derived stem cells and lung and mammary tissues. (B) NEAT1 expression is increased by overexpression (OE) of miR-140 in ATCC (human lung fibroblasts) and 3T3-L1 (preadipocytes) cells. (C) Adipocyte-derived stem cells obtained from WT and miR-140 KO mice were stained with NEAT1 probe and antidigoxigenin (anti-DIG) antibodies (for miR-140) and visualized by fluorescence. Shown are expression levels of m140 and NEAT1 in WT and miR-140 KO adipocyte-derived stem cells. (D) 3T3-L1 cells were transfected with either pCAG-GFP (control vector) or miR-140 overexpression vector. The transfected cells were fixed and analyzed for changes in the expression levels of NEAT1 through in situ hybridization of NEAT1 and miR-140 probes. In situ hybridization of miR-140 (5′ DIG tagged; identified using anti-DIG Cy3-conjugated antibody) and NEAT1 probes (FITC conjugated). Shown are expression levels of miR-140 and NEAT1 in WT and miR-140 (m140) in control and miR-140-overexpressing 3T3-L1 cells. (E) Demonstration of NEAT1 stability in miR-140-overexpressing 3T3-L1 cells. The data represent means and SE (n = 3). *, P < 0.05; **, P < 0.01.
FIG 5
Effects of NEAT1 overexpression on the process of differentiation. (A) Demonstration of numbers and sizes of spheres with adipocyte-derived stem cells of WT and miR-140 KO mice. (B) Quantification of spheres measuring more than 100 μm. (C) Fold change of OCT-4 levels in the spheres of WT and miR-140 KO cells by RT-PCR. (D) NEAT1 overexpression in miR-140 KO adipocyte-derived stem cells resulted in an increase in differentiation. (E) Confirmation of NEAT1 overexpression by RT-PCR. (F) Representation of the amount of Oil Red dye in lipid droplets by ImageJ analysis. (G and H) Expression levels of CEBP/α and PPARγ in vector (control) and NEAT1-overexpressing miR-140 KO adipocyte-derived stem cells. The data represent means and SE (n = 3). *, P < 0.05; **, P < 0.01. Scale bars (A and D), 100 μm.
Similar articles
- Overexpression of lncRNA-NEF regulates the miR-155/PTEN axis to inhibit adipogenesis and promote osteogenesis.
Ming Y, Liu ZP. Ming Y, et al. Kaohsiung J Med Sci. 2021 Nov;37(11):930-939. doi: 10.1002/kjm2.12423. Epub 2021 Aug 12. Kaohsiung J Med Sci. 2021. PMID: 34382731 Free PMC article. - The novel long noncoding RNA lncRNA-Adi regulates adipogenesis.
Chen Y, Li K, Zhang X, Chen J, Li M, Liu L. Chen Y, et al. Stem Cells Transl Med. 2020 Sep;9(9):1053-1067. doi: 10.1002/sctm.19-0438. Epub 2020 May 1. Stem Cells Transl Med. 2020. PMID: 32356938 Free PMC article. - Obesity-Associated MiR-342-3p Promotes Adipogenesis of Mesenchymal Stem Cells by Suppressing CtBP2 and Releasing C/EBPα from CtBP2 Binding.
Wang L, Xu L, Xu M, Liu G, Xing J, Sun C, Ding H. Wang L, et al. Cell Physiol Biochem. 2015;35(6):2285-98. doi: 10.1159/000374032. Epub 2015 Apr 13. Cell Physiol Biochem. 2015. PMID: 25895816 - Long noncoding RNAs in regulating adipogenesis: new RNAs shed lights on obesity.
Wei S, Du M, Jiang Z, Hausman GJ, Zhang L, Dodson MV. Wei S, et al. Cell Mol Life Sci. 2016 May;73(10):2079-87. doi: 10.1007/s00018-016-2169-2. Epub 2016 Mar 4. Cell Mol Life Sci. 2016. PMID: 26943803 Free PMC article. Review. - microRNAs in the regulation of adipogenesis and obesity.
McGregor RA, Choi MS. McGregor RA, et al. Curr Mol Med. 2011 Jun;11(4):304-16. doi: 10.2174/156652411795677990. Curr Mol Med. 2011. PMID: 21506921 Free PMC article. Review.
Cited by
- Long noncoding RNA LYPLAL1-AS1 regulates adipogenic differentiation of human mesenchymal stem cells by targeting desmoplakin and inhibiting the Wnt/β-catenin pathway.
Yang Y, Fan J, Xu H, Fan L, Deng L, Li J, Li D, Li H, Zhang F, Zhao RC. Yang Y, et al. Cell Death Discov. 2021 May 15;7(1):105. doi: 10.1038/s41420-021-00500-5. Cell Death Discov. 2021. PMID: 33993187 Free PMC article. - Dynamic Expression Profiles of Circular RNAs during Brown to White Adipose Tissue Transformation in Goats (Capra hircus).
Zhang X, Zhan S, Yang S, Zhong T, Guo J, Cao J, Wang Y, Li L, Zhang H, Wang L. Zhang X, et al. Animals (Basel). 2021 May 10;11(5):1351. doi: 10.3390/ani11051351. Animals (Basel). 2021. PMID: 34068539 Free PMC article. - Long non-coding RNA TCONS_00041960 enhances osteogenesis and inhibits adipogenesis of rat bone marrow mesenchymal stem cell by targeting miR-204-5p and miR-125a-3p.
Shang G, Wang Y, Xu Y, Zhang S, Sun X, Guan H, Zhao X, Wang Y, Li Y, Zhao G. Shang G, et al. J Cell Physiol. 2018 Aug;233(8):6041-6051. doi: 10.1002/jcp.26424. Epub 2018 Feb 28. J Cell Physiol. 2018. PMID: 29319166 Free PMC article. - Human Long Noncoding RNA Regulation of Stem Cell Potency and Differentiation.
Lee S, Seo HH, Lee CY, Lee J, Shin S, Kim SW, Lim S, Hwang KC. Lee S, et al. Stem Cells Int. 2017;2017:6374504. doi: 10.1155/2017/6374504. Epub 2017 Aug 30. Stem Cells Int. 2017. PMID: 28951743 Free PMC article. Review. - Unraveling NEAT1's complex role in lung cancer biology: a comprehensive review.
Hussain MS, Afzal O, Gupta G, Goyal A, Almalki WH, Kazmi I, Alzarea SI, Alfawaz Altamimi AS, Kukreti N, Chakraborty A, Singh SK, Dua K. Hussain MS, et al. EXCLI J. 2024 Jan 3;23:34-52. doi: 10.17179/excli2023-6553. eCollection 2024. EXCLI J. 2024. PMID: 38343745 Free PMC article. Review.
References
- Dirat B, Bochet L, Dabek M, Daviaud D, Dauvillier S, Majed B, Wang Y-Y, Meulle A, Salles B, Le Gonidec S, Garrido I, Escourrou G, Valet P, Muller C. 2011. Cancer-associated adipocytes exhibit an activated phenotype and contribute to breast cancer invasion. Cancer Res 71:2455–2465. doi:10.1158/0008-5472.CAN-10-3323. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 AR050631/AR/NIAMS NIH HHS/United States
- R01 AR065379/AR/NIAMS NIH HHS/United States
- R01 CA163820/CA/NCI NIH HHS/United States
- AR065379/AR/NIAMS NIH HHS/United States
- CA163820A1/CA/NCI NIH HHS/United States
- CA157779A1/CA/NCI NIH HHS/United States
- AR050631/AR/NIAMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Molecular Biology Databases