FlyBase: establishing a Gene Group resource for Drosophila melanogaster - PubMed (original) (raw)
. 2016 Jan 4;44(D1):D786-92.
doi: 10.1093/nar/gkv1046. Epub 2015 Oct 13.
Collaborators, Affiliations
- PMID: 26467478
- PMCID: PMC4702782
- DOI: 10.1093/nar/gkv1046
FlyBase: establishing a Gene Group resource for Drosophila melanogaster
Helen Attrill et al. Nucleic Acids Res. 2016.
Abstract
Many publications describe sets of genes or gene products that share a common biology. For example, genome-wide studies and phylogenetic analyses identify genes related in sequence; high-throughput genetic and molecular screens reveal functionally related gene products; and advanced proteomic methods can determine the subunit composition of multi-protein complexes. It is useful for such gene collections to be presented as discrete lists within the appropriate Model Organism Database (MOD) so that researchers can readily access these data alongside other relevant information. To this end, FlyBase (flybase.org), the MOD for Drosophila melanogaster, has established a 'Gene Group' resource: high-quality sets of genes derived from the published literature and organized into individual report pages. To facilitate further analyses, Gene Group Reports also include convenient download and analysis options, together with links to equivalent gene groups at other databases. This new resource will enable researchers with diverse backgrounds and interests to easily view and analyse acknowledged D. melanogaster gene sets and compare them with those of other species.
© The Author(s) 2015. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures
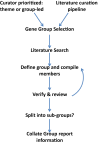
Figure 1.
Gene Group Curation Strategy. A generalized scheme detailing the workflow for producing Gene Groups in FlyBase. See text for details.
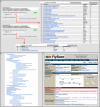
Figure 2.
Finding Gene Groups. (A) The QuickSearch ‘Gene Groups’ tab can be searched using a symbol/name of a group (a search for ‘GPCR’ returns 29 hits that contain ‘GPCR’ in the text of the Report) or a member gene (a search for ‘_ninaE_’ yields 3 hits from the rhodopsin GPCR hierarchy). (B) Clicking on the ‘browse’ link in the ‘Gene Groups’ tab of QuickSearch takes the user to an alphabetical, nested list of all FlyBase Gene Groups. The section shown focuses on the GPCR hierarchy. (C) Gene Group membership is indicated within the ‘Gene Group Membership (FlyBase)’ field (red box) of the ‘Families, Domains and Molecular Function’ section of the Gene Report. In this example, the upper part of the ninaE Gene Report page is shown and the RHODOPSINS Gene Group is displayed.
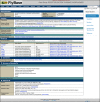
Figure 3.
The Gene Group Report. The example shown is for the RECEPTOR PROTEIN TYROSINE PHOSPHATASES group. This group has one immediate parent group (PROTEIN TYROSINE PHOSPHATASES), shown in the ‘Related Gene Groups’ subsection. See text for details.
Similar articles
- Using FlyBase to Find Functionally Related Drosophila Genes.
Rey AJ, Attrill H, Marygold SJ; FlyBase Consortium. Rey AJ, et al. Methods Mol Biol. 2018;1757:493-512. doi: 10.1007/978-1-4939-7737-6_16. Methods Mol Biol. 2018. PMID: 29761468 Free PMC article. - Using FlyBase, a Database of Drosophila Genes and Genomes.
Marygold SJ, Crosby MA, Goodman JL; FlyBase Consortium. Marygold SJ, et al. Methods Mol Biol. 2016;1478:1-31. doi: 10.1007/978-1-4939-6371-3_1. Methods Mol Biol. 2016. PMID: 27730573 Free PMC article. Review. - FlyBase : a database for the Drosophila research community.
Drysdale R; FlyBase Consortium. Drysdale R, et al. Methods Mol Biol. 2008;420:45-59. doi: 10.1007/978-1-59745-583-1_3. Methods Mol Biol. 2008. PMID: 18641940 Review. - FlyBase: integration and improvements to query tools.
Wilson RJ, Goodman JL, Strelets VB; FlyBase Consortium. Wilson RJ, et al. Nucleic Acids Res. 2008 Jan;36(Database issue):D588-93. doi: 10.1093/nar/gkm930. Epub 2007 Dec 26. Nucleic Acids Res. 2008. PMID: 18160408 Free PMC article. - FlyBase: updates to the Drosophila genes and genomes database.
Öztürk-Çolak A, Marygold SJ, Antonazzo G, Attrill H, Goutte-Gattat D, Jenkins VK, Matthews BB, Millburn G, Dos Santos G, Tabone CJ; FlyBase Consortium. Öztürk-Çolak A, et al. Genetics. 2024 May 7;227(1):iyad211. doi: 10.1093/genetics/iyad211. Genetics. 2024. PMID: 38301657 Free PMC article.
Cited by
- Loss of Drosophila nucleostemin 2 (NS2) blocks nucleolar release of the 60S subunit leading to ribosome stress.
Wang Y, DiMario P. Wang Y, et al. Chromosoma. 2017 Jun;126(3):375-388. doi: 10.1007/s00412-016-0597-2. Epub 2016 May 6. Chromosoma. 2017. PMID: 27150106 - Partial Functional Diversification of Drosophila melanogaster Septin Genes Sep2 and Sep5.
O'Neill RS, Clark DV. O'Neill RS, et al. G3 (Bethesda). 2016 Jul 7;6(7):1947-57. doi: 10.1534/g3.116.028886. G3 (Bethesda). 2016. PMID: 27172205 Free PMC article. - The translation factors of Drosophila melanogaster.
Marygold SJ, Attrill H, Lasko P. Marygold SJ, et al. Fly (Austin). 2017 Jan 2;11(1):65-74. doi: 10.1080/19336934.2016.1220464. Epub 2016 Aug 5. Fly (Austin). 2017. PMID: 27494710 Free PMC article. - Ohgata, the Single Drosophila Ortholog of Human Cereblon, Regulates Insulin Signaling-dependent Organismic Growth.
Wakabayashi S, Sawamura N, Voelzmann A, Broemer M, Asahi T, Hoch M. Wakabayashi S, et al. J Biol Chem. 2016 Nov 25;291(48):25120-25132. doi: 10.1074/jbc.M116.757823. Epub 2016 Oct 4. J Biol Chem. 2016. PMID: 27702999 Free PMC article. - Drosophila Ringmaker regulates microtubule stabilization and axonal extension during embryonic development.
Mino RE, Rogers SL, Risinger AL, Rohena C, Banerjee S, Bhat MA. Mino RE, et al. J Cell Sci. 2016 Sep 1;129(17):3282-94. doi: 10.1242/jcs.187294. Epub 2016 Jul 15. J Cell Sci. 2016. PMID: 27422099 Free PMC article.
References
- Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. - PubMed
- Gray K.A., Yates B., Seal R.L., Wright M.W., Bruford E.A. Genenames.org: the HGNC resources in 2015. Nucleic Acids Res. 2015;43:D1079–1085. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases