Genetic Variability and Structuring of Arctic Charr (Salvelinus alpinus) Populations in Northern Fennoscandia - PubMed (original) (raw)
Genetic Variability and Structuring of Arctic Charr (Salvelinus alpinus) Populations in Northern Fennoscandia
Takahito Shikano et al. PLoS One. 2015.
Abstract
Variation in presumably neutral genetic markers can inform us about evolvability, historical effective population sizes and phylogeographic history of contemporary populations. We studied genetic variability in 15 microsatellite loci in six native landlocked Arctic charr (Salvelinus alpinus) populations in northern Fennoscandia, where this species is considered near threatened. We discovered that all populations were genetically highly (mean FST ≈ 0.26) differentiated and isolated from each other. Evidence was found for historical, but not for recent population size bottlenecks. Estimates of contemporary effective population size (Ne) ranged from seven to 228 and were significantly correlated with those of historical Ne but not with lake size. A census size (NC) was estimated to be approximately 300 individuals in a pond (0.14 ha), which exhibited the smallest Ne (i.e. Ne/NC = 0.02). Genetic variability in this pond and a connected lake is severely reduced, and both genetic and empirical estimates of migration rates indicate a lack of gene flow between them. Hence, albeit currently thriving, some northern Fennoscandian populations appear to be vulnerable to further loss of genetic variability and are likely to have limited capacity to adapt if selection pressures change.
Conflict of interest statement
Competing Interests: The authors have declared that no competing interests exist.
Figures
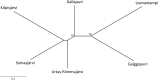
Fig 1. Sampling locations of the six Arctic charr populations.
The current situation is presented in black, in which Lakes Kilpisjärvi, Urtas-Riimmajärvi and Somasjärvi belong to the watercourse draining into the Baltic Sea and the other lakes belong to the watercourse draining into the Atlantic Ocean. Gray shading indicates the historical (ca. 10 000 years ago) situation, when all lakes were involved in two separate watercourses draining into the Atlantic Ocean. All studied populations are landlocked at the present day.
Fig 2. An unrooted neighbor-joining tree based on D A distances among the six Arctic charr populations.
Bootstrap support (>50%) is given at each node.
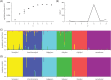
Fig 3. Bayesian clustering analyses for the six Arctic charr populations based on STRUCTURE and BAPS.
(A) Likelihood estimates for K = 1–9 in STRUCTURE. (B) Estimated delta K for K = 2–8 in STRUCTURE. (C) Individual assignment at K = 6 in STRUCTURE. (D) Individual assignment at K = 6 in BAPS. Each individual is shown in a vertical bar in the same sequence (C and D).
Similar articles
- Phylogeography and sympatric differentiation of the Arctic charr Salvelinus alpinus (L.) complex in Siberia as revealed by mtDNA sequence analysis.
Alekseyev SS, Bajno R, Gordeeva NV, Reist JD, Power M, Kirillov AF, Samusenok VP, Matveev AN. Alekseyev SS, et al. J Fish Biol. 2009 Aug;75(2):368-92. doi: 10.1111/j.1095-8649.2009.02331.x. J Fish Biol. 2009. PMID: 20738544 - Holarctic phylogeography of Arctic charr (Salvelinus alpinus L.) inferred from mitochondrial DNA sequences.
Brunner PC, Douglas MR, Osinov A, Wilson CC, Bernatchez L. Brunner PC, et al. Evolution. 2001 Mar;55(3):573-86. doi: 10.1554/0014-3820(2001)055[0573:hpoacs]2.0.co;2. Evolution. 2001. PMID: 11327164 - Population genetic structure of Arctic charr, Salvelinus alpinus from northwest Europe on large and small spatial scales.
Wilson AJ, Gíslason D, Skúlason S, Snorrason SS, Adams CE, Alexander G, Danzmann RG, Ferguson MM. Wilson AJ, et al. Mol Ecol. 2004 May;13(5):1129-42. doi: 10.1111/j.1365-294X.2004.02149.x. Mol Ecol. 2004. PMID: 15078451 - Evolution of adaptive diversity and genetic connectivity in Arctic charr (Salvelinus alpinus) in Iceland.
Kapralova KH, Morrissey MB, Kristjánsson BK, Olafsdóttir GÁ, Snorrason SS, Ferguson MM. Kapralova KH, et al. Heredity (Edinb). 2011 Mar;106(3):472-87. doi: 10.1038/hdy.2010.161. Epub 2011 Jan 12. Heredity (Edinb). 2011. PMID: 21224880 Free PMC article.
Cited by
- Characterizing neutral and adaptive genomic differentiation in a changing climate: The most northerly freshwater fish as a model.
O'Malley KG, Vaux F, Black AN. O'Malley KG, et al. Ecol Evol. 2019 Jan 15;9(4):2004-2017. doi: 10.1002/ece3.4891. eCollection 2019 Feb. Ecol Evol. 2019. PMID: 30847088 Free PMC article. - Fast and accurate population admixture inference from genotype data from a few microsatellites to millions of SNPs.
Wang J. Wang J. Heredity (Edinb). 2022 Aug;129(2):79-92. doi: 10.1038/s41437-022-00535-z. Epub 2022 May 4. Heredity (Edinb). 2022. PMID: 35508539 Free PMC article. - Landscape genetic structure of Scirpus mariqueter reveals a putatively adaptive differentiation under strong gene flow in estuaries.
Yang M, Xu C, Duchesne P, Ma Q, Yin G, Fang Y, Lu F, Zhang W. Yang M, et al. Ecol Evol. 2019 Feb 28;9(6):3059-3074. doi: 10.1002/ece3.4793. eCollection 2019 Mar. Ecol Evol. 2019. PMID: 30962881 Free PMC article.
References
- DeWoody JA, Avise JC (2000) Microsatellite variation in marine, freshwater and anadromous fishes compared with other animals. J Fish Biol 56: 461–473.
- Merilä J (2014) Lakes and ponds as model systems to study convergent and parallel evolution. J. Limnol 73: 33–45.
- Frankham R, Ballou JD, Briscoe DA (2002) Introduction to Conservation Genetics. Cambridge: Cambridge University Press.
- Hewitt G (2000) The genetic legacy of the Quaternary ice ages. Nature 405: 907–913. - PubMed
Publication types
MeSH terms
Grants and funding
This research was supported by grants from Academy of Finland (to JM) and Ministry of Agriculture and Forestry (to KKK).
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous