Divergent responses of viral and bacterial communities in the gut microbiome to dietary disturbances in mice - PubMed (original) (raw)
Divergent responses of viral and bacterial communities in the gut microbiome to dietary disturbances in mice
Adina Howe et al. ISME J. 2016 May.
Abstract
To improve our understanding of the stability of mammalian intestinal communities, we characterized the responses of both bacterial and viral communities in murine fecal samples to dietary changes between high- and low-fat (LF) diets. Targeted DNA extraction methods for bacteria, virus-like particles and induced prophages were used to generate bacterial and viral metagenomes as well as 16S ribosomal RNA amplicons. Gut microbiome communities from two cohorts of C57BL/6 mice were characterized in a 6-week diet perturbation study in response to high fiber, LF and high-refined sugar, milkfat (MF) diets. The resulting metagenomes from induced bacterial prophages and extracellular viruses showed significant overlap, supporting a largely temperate viral lifestyle within these gut microbiomes. The resistance of baseline communities to dietary disturbances was evaluated, and we observed contrasting responses of baseline LF and MF bacterial and viral communities. In contrast to baseline LF viral communities and bacterial communities in both diet treatments, baseline MF viral communities were sensitive to dietary disturbances as reflected in their non-recovery during the washout period. The contrasting responses of bacterial and viral communities suggest that these communities can respond to perturbations independently of each other and highlight the potentially unique role of viruses in gut health.
Figures
Figure 1
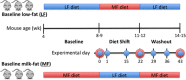
Study design of two mouse cohorts (_n_=3) with varying diet treatments (baseline low-fat (LF) and high-milkfat (MF) diets). Weekly fecal samples were collected for microbial community composition (16S rRNA amplicon analysis, blue) and function (metagenomic sequencing, red).
Figure 2
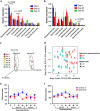
Standardized relative abundances (abundances standardized by total number of reads for each sample) of annotated contigs, which could be assigned to (a) taxa or (b) functions by MG-RAST M5NR SEED database.
Figure 3
Analysis of bacterial community structure through 16S rRNA genes. Mean relative abundances of order level taxa in fecal bacterial communities present at >1% in (a) baseline LF and (b) baseline MF mice. Data are shown as mean±s.e.m.; _P_-values (ANOVA) as shown on graph. (c) Principal coordinate analysis plot of the bacterial community structure based on Bray–Curtis distances. Symbols represent individual mice, coded by color to represent collection day. (d) Principal coordinate analysis of second and third principal coordinate axes, based on Bray–Curtis distances. Colored symbols are the diet the mice were consuming on each experimental day, denoted as the number below each symbol. Changes in alpha diversity of fecal bacterial communities during dietary perturbation and washout: (e) Shannon diversity index values and (f) number of observed species. Data are shown as mean±s.e.m., _P_-values are shown *P<0.05, **P<0.01, ***P<0.001, ****P<0.0001 comparing the means of both treatment groups at each time point; ANOVA, Bonferroni's multiple comparison test.
Figure 4
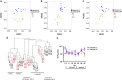
NMDS of Bray–Curtis distances of sequence abundances in (a) BAC, (b) VLP and (c) IND metagenomes. Marker colors represent different diet treatments (orange=baseline MF, blue=baseline LF) and marker shapes indicate day of sample. (d) Average similarity hierarchical clustering of BAC, IND and VLP metagenomes on Day 0 (black, baseline) and Day 43 (red, washout) for baseline LF and MF samples, based on Bray–Curtis distances of median bp coverage of sequences. (e) Direct counts of VLPs from VLP fraction. Mean VLP counts±s.e.m. are shown. *P<0.05, **P<0.01.
Figure 5
Co-occurrence network representing relationships between significant contigs identified in baseline MF virus fractions (see Supplementary Table S7) and 16S rRNA amplicons and BAC metagenome contigs. Taxonomic orders of sequences that share the most similarity in the MG-RAST database are shown for contigs in the baseline MF VLP (a) and IND (b) samples. Three separated co-occurrence networks have been labeled and identified as network modules.
Similar articles
- Fecal Viral Community Responses to High-Fat Diet in Mice.
Schulfer A, Santiago-Rodriguez TM, Ly M, Borin JM, Chopyk J, Blaser MJ, Pride DT. Schulfer A, et al. mSphere. 2020 Feb 26;5(1):e00833-19. doi: 10.1128/mSphere.00833-19. mSphere. 2020. PMID: 32102942 Free PMC article. - Adverse effect of early-life high-fat/high-carbohydrate ("Western") diet on bacterial community in the distal bowel of mice.
Villamil SI, Huerlimann R, Morianos C, Sarnyai Z, Maes GE. Villamil SI, et al. Nutr Res. 2018 Feb;50:25-36. doi: 10.1016/j.nutres.2017.11.008. Epub 2017 Dec 6. Nutr Res. 2018. PMID: 29540269 - Dietary fiber intake impacts gut bacterial and viral populations in a hypertensive mouse model.
Avellaneda-Franco L, Xie L, Nakai M, Barr JJ, Marques FZ. Avellaneda-Franco L, et al. Gut Microbes. 2024 Jan-Dec;16(1):2407047. doi: 10.1080/19490976.2024.2407047. Epub 2024 Sep 28. Gut Microbes. 2024. PMID: 39340212 Free PMC article. - Experimental Manipulation Shows a Greater Influence of Population than Dietary Perturbation on the Microbiome of Tyrophagus putrescentiae.
Erban T, Ledvinka O, Nesvorna M, Hubert J. Erban T, et al. Appl Environ Microbiol. 2017 Apr 17;83(9):e00128-17. doi: 10.1128/AEM.00128-17. Print 2017 May 1. Appl Environ Microbiol. 2017. PMID: 28235879 Free PMC article. - Current state of knowledge: the canine gastrointestinal microbiome.
Hooda S, Minamoto Y, Suchodolski JS, Swanson KS. Hooda S, et al. Anim Health Res Rev. 2012 Jun;13(1):78-88. doi: 10.1017/S1466252312000059. Epub 2012 May 30. Anim Health Res Rev. 2012. PMID: 22647637 Review.
Cited by
- Bacteriophages: Uncharacterized and Dynamic Regulators of the Immune System.
Sinha A, Maurice CF. Sinha A, et al. Mediators Inflamm. 2019 Sep 8;2019:3730519. doi: 10.1155/2019/3730519. eCollection 2019. Mediators Inflamm. 2019. PMID: 31582898 Free PMC article. Review. - Characterization of Phietavirus Henu 2 in the virome of individuals with acute gastroenteritis.
do Socorro Fôro Ramos E, Bahia SL, de Oliveira Ribeiro G, Villanova F, de Pádua Milagres FA, Brustulin R, Pandey RP, Deng X, Delwart E, da Costa AC, Leal É. do Socorro Fôro Ramos E, et al. Virus Genes. 2023 Jun;59(3):464-472. doi: 10.1007/s11262-023-01990-4. Epub 2023 Apr 1. Virus Genes. 2023. PMID: 37004601 Review. - Metabolic Modeling Elucidates the Transactions in the Rumen Microbiome and the Shifts Upon Virome Interactions.
Islam MM, Fernando SC, Saha R. Islam MM, et al. Front Microbiol. 2019 Oct 22;10:2412. doi: 10.3389/fmicb.2019.02412. eCollection 2019. Front Microbiol. 2019. PMID: 31866953 Free PMC article. - Integrative Longitudinal Analysis of Metabolic Phenotype and Microbiota Changes During the Development of Obesity.
Higgins KV, Woodie LN, Hallowell H, Greene MW, Schwartz EH. Higgins KV, et al. Front Cell Infect Microbiol. 2021 Aug 3;11:671926. doi: 10.3389/fcimb.2021.671926. eCollection 2021. Front Cell Infect Microbiol. 2021. PMID: 34414128 Free PMC article. - Western diets, gut dysbiosis, and metabolic diseases: Are they linked?
Martinez KB, Leone V, Chang EB. Martinez KB, et al. Gut Microbes. 2017 Mar 4;8(2):130-142. doi: 10.1080/19490976.2016.1270811. Epub 2017 Jan 6. Gut Microbes. 2017. PMID: 28059614 Free PMC article. Review.
References
- Breitbart M, Haynes M, Kelley S, Angly F, Edwards RA, Felts B et al. (2008). Viral diversity and dynamics in an infant gut. Res Microbiol 159: 367–373. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- P30 DK042086/DK/NIDDK NIH HHS/United States
- T32 DK007074/DK/NIDDK NIH HHS/United States
- DK07074/DK/NIDDK NIH HHS/United States
- DK097268/DK/NIDDK NIH HHS/United States
- R01 DK097268/DK/NIDDK NIH HHS/United States
- P30 DK42086/DK/NIDDK NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources