Within-host microevolution of Pseudomonas aeruginosa in Italian cystic fibrosis patients - PubMed (original) (raw)
Case Reports
Within-host microevolution of Pseudomonas aeruginosa in Italian cystic fibrosis patients
Rasmus Lykke Marvig et al. BMC Microbiol. 2015.
Abstract
Background: Chronic infection with Pseudomonas aeruginosa is a major cause of morbidity and mortality in cystic fibrosis (CF) patients, and a more complete understanding of P. aeruginosa within-host genomic evolution, transmission, and population genomics may provide a basis for improving intervention strategies. Here, we report the first genomic analysis of P. aeruginosa isolates sampled from Italian CF patients.
Results: By genome sequencing of 26 isolates sampled over 19 years from four patients, we elucidated the within-host evolution of clonal lineages in each individual patient. Many of the identified mutations were located in pathoadaptive genes previously associated with host adaptation, and we correlated mutations with changes in CF-relevant phenotypes such as antibiotic resistance. In addition, the genomic analysis revealed that three patients shared the same clone. Furthermore, we compared the genomes of the Italian CF isolates to a panel of genome sequenced strains of P. aeruginosa from other countries. Isolates from two of the Italian lineages belonged to clonal complexes of P. aeruginosa that have previously been identified in Danish CF patients, and our genomic comparison showed that clonal isolates from the same country may be more distantly related than clonal isolates from different countries.
Conclusions: This is the first whole-genome analysis of P. aeruginosa isolated from Italian CF patients, and together with both phenotypic and clinical information this dataset facilitates a more detailed understanding of P. aeruginosa within-host genomic evolution, transmission, and population genomics. We conclude that the evolution of the Italian lineages resembles what has been found in other countries.
Figures
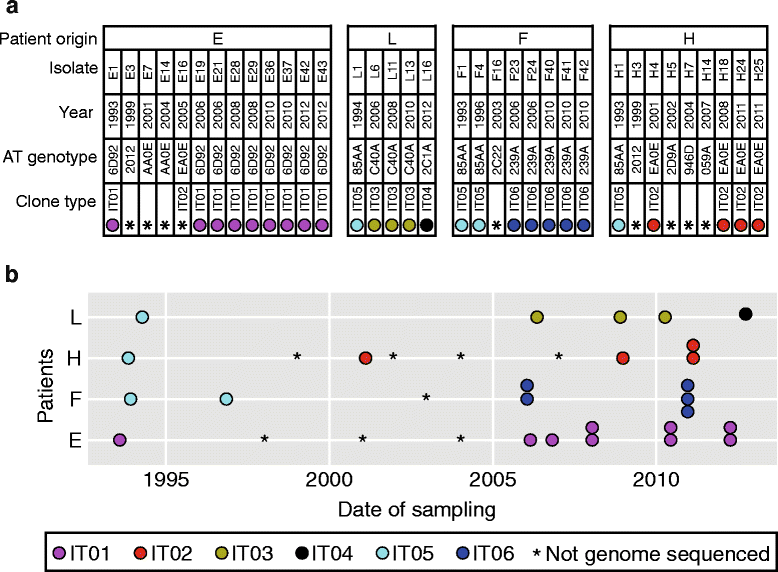
Fig. 1
Collection of Pseudomonas aeruginosa isolates. a Information about P. aeruginosa isolates collected from patients E, L, F, and H. AT genotype refers to the genotype code determined by ArrayTube multimarker microarray. b Longitudinal overview of samples
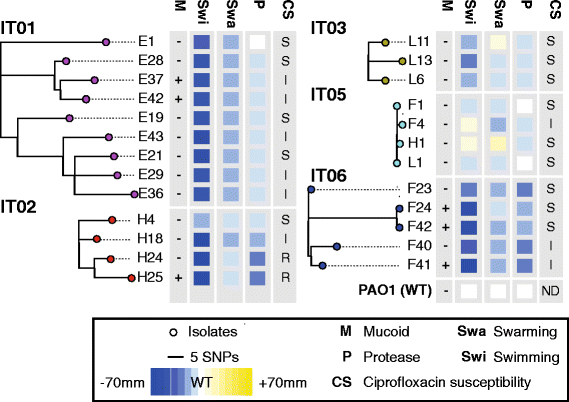
Fig. 2
Phylogeny and selected phenotypes of Pseudomonas aeruginosa lineages IT01, IT02, IT03, IT05, and IT06. Maximum-parsimonious phylogenetic trees were constructed by PAUP* (Methods). Size of swimming, swarming, and protease clearing zones relative to reference strain PAO1 are indicated by colors. Presence of mucoid phenotype is denoted by ‘+’. Ciprofloxacin susceptibility is indicated by ‘S’ (>21 mm clearing zone), ‘I’ (16–20 mm clearing zone), or ´R´ (no clearing zone)
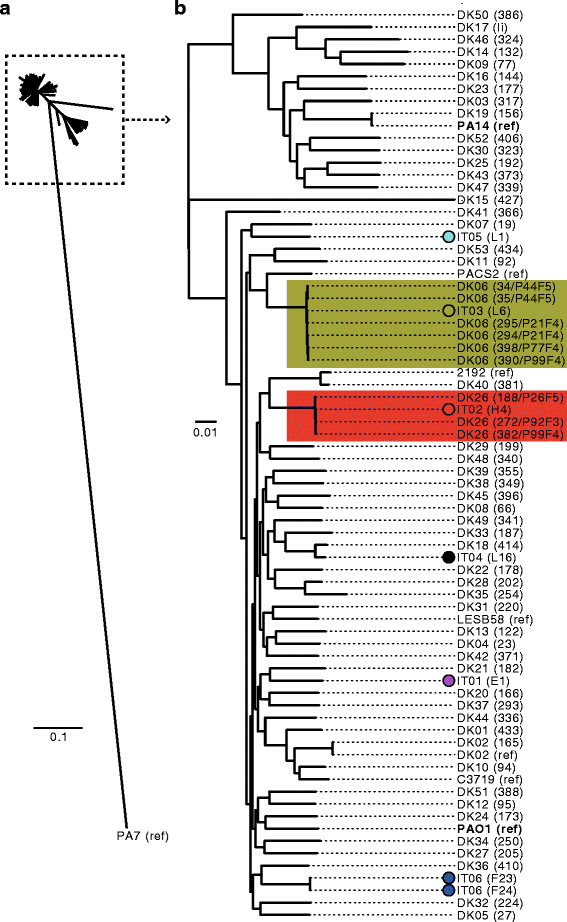
Fig. 3
Phylogenetic tree based on core-genome alignment of different Pseudomonas aeruginosa strains. Genomes of the earliest isolate(s) of each of the lineages IT01-IT06 were aligned against a panel of 60 genome sequences of other strains of P. aeruginosa. a Tree showing the phylogeny of all isolates. b Subset of tree showing phylogeny of all isolates, except the outlier isolate PA7. Strains are either named after their lineage (IT01-IT06 or DK01-DK53) followed by the name of the specific isolate in parenthesis, or by the name of the reference strains (strains with completed genome sequences) followed by ‘ref’ in parenthesis. Phylogenetic tree was constructed using Harvest [28]
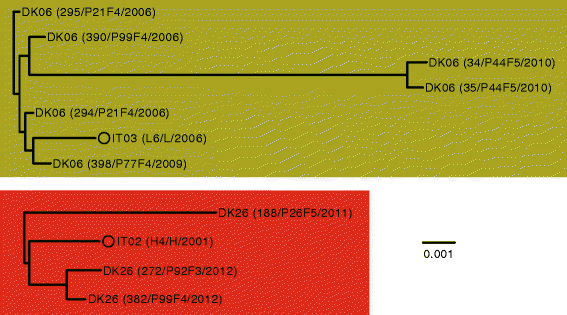
Fig. 4
Phylogenetic trees based on core-genome alignment of Pseudomonas aeruginosa strains of the IT02/DK26 and IT03/DK06 clonal complexes. Phylogenetic trees were constructed using Harvest [28]. Name, patient origin, and year of isolation are shown for each isolate in parenthesis
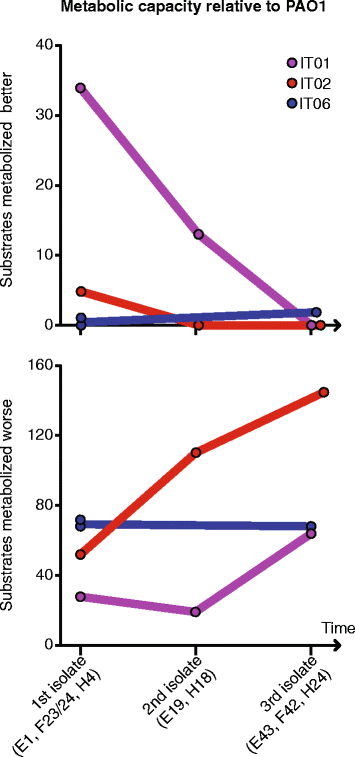
Fig. 5
Changes in metabolic capacities over time relative to Pseudomonas aeruginosa reference strain PAO1. Number of substrates metabolized better or worse than PAO1 was measured using Biolog Phenotype MicroArrays (Biolog, Hayward, CA)
Similar articles
- Evolutionary insight from whole-genome sequencing of Pseudomonas aeruginosa from cystic fibrosis patients.
Marvig RL, Sommer LM, Jelsbak L, Molin S, Johansen HK. Marvig RL, et al. Future Microbiol. 2015;10(4):599-611. doi: 10.2217/fmb.15.3. Future Microbiol. 2015. PMID: 25865196 Review. - Genotypic and phenotypic relatedness of Pseudomonas aeruginosa isolates among the major cystic fibrosis patient cohort in Italy.
Cigana C, Melotti P, Baldan R, Pedretti E, Pintani E, Iansa P, De Fino I, Favari F, Bergamini G, Tridello G, Cirillo DM, Assael BM, Bragonzi A. Cigana C, et al. BMC Microbiol. 2016 Jul 11;16(1):142. doi: 10.1186/s12866-016-0760-1. BMC Microbiol. 2016. PMID: 27400750 Free PMC article. - Selective Sweeps and Parallel Pathoadaptation Drive Pseudomonas aeruginosa Evolution in the Cystic Fibrosis Lung.
Diaz Caballero J, Clark ST, Coburn B, Zhang Y, Wang PW, Donaldson SL, Tullis DE, Yau YC, Waters VJ, Hwang DM, Guttman DS. Diaz Caballero J, et al. mBio. 2015 Sep 1;6(5):e00981-15. doi: 10.1128/mBio.00981-15. mBio. 2015. PMID: 26330513 Free PMC article. - Convergent evolution and adaptation of Pseudomonas aeruginosa within patients with cystic fibrosis.
Marvig RL, Sommer LM, Molin S, Johansen HK. Marvig RL, et al. Nat Genet. 2015 Jan;47(1):57-64. doi: 10.1038/ng.3148. Epub 2014 Nov 17. Nat Genet. 2015. PMID: 25401299
Cited by
- Omics-based tracking of Pseudomonas aeruginosa persistence in "eradicated" cystic fibrosis patients.
Bartell JA, Sommer LM, Marvig RL, Skov M, Pressler T, Molin S, Johansen HK. Bartell JA, et al. Eur Respir J. 2021 Apr 8;57(4):2000512. doi: 10.1183/13993003.00512-2020. Print 2021 Apr. Eur Respir J. 2021. PMID: 33093121 Free PMC article. - Convergent Metabolic Specialization through Distinct Evolutionary Paths in Pseudomonas aeruginosa.
La Rosa R, Johansen HK, Molin S. La Rosa R, et al. mBio. 2018 Apr 10;9(2):e00269-18. doi: 10.1128/mBio.00269-18. mBio. 2018. PMID: 29636437 Free PMC article. - Forecasting of phenotypic and genetic outcomes of experimental evolution in Pseudomonas protegens.
Pentz JT, Lind PA. Pentz JT, et al. PLoS Genet. 2021 Aug 5;17(8):e1009722. doi: 10.1371/journal.pgen.1009722. eCollection 2021 Aug. PLoS Genet. 2021. PMID: 34351900 Free PMC article. - From genotype to phenotype: adaptations of Pseudomonas aeruginosa to the cystic fibrosis environment.
Camus L, Vandenesch F, Moreau K. Camus L, et al. Microb Genom. 2021 Mar;7(3):mgen000513. doi: 10.1099/mgen.0.000513. Epub 2021 Feb 2. Microb Genom. 2021. PMID: 33529147 Free PMC article. Review. - Pseudomonas aeruginosa faces a fitness trade-off between mucosal colonization and antibiotic tolerance during airway infection.
Meirelles LA, Vayena E, Debache A, Schmidt E, Rossy T, Distler T, Hatzimanikatis V, Persat A. Meirelles LA, et al. Nat Microbiol. 2024 Dec;9(12):3284-3303. doi: 10.1038/s41564-024-01842-3. Epub 2024 Oct 25. Nat Microbiol. 2024. PMID: 39455898
References
- Marvig RL, Sommer LM, Jelsbak L, Molin S, Johansen HK. Evolutionary insight from whole-genome sequencing of Pseudomonas aeruginosa from cystic fibrosis patients. Future Microbiol. 2015;10. - PubMed
- Feliziani S, Marvig RL, Lujan AM, Moyano AJ, Di Rienzo JA, Krogh Johansen H, et al. Coexistence and Within-Host Evolution of Diversified Lineages of Hypermutable Pseudomonas aeruginosa in Long-term Cystic Fibrosis Infections. PLoS Genet. 2014;10 doi: 10.1371/journal.pgen.1004651. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical