Perinatal deiodinase 2 expression in hepatocytes defines epigenetic susceptibility to liver steatosis and obesity - PubMed (original) (raw)
. 2015 Nov 10;112(45):14018-23.
doi: 10.1073/pnas.1508943112. Epub 2015 Oct 27.
Gustavo W Fernandes 2, Elizabeth A McAninch 1, Barbara M L C Bocco 2, Sherine M Abdalla 3, Miriam O Ribeiro 4, Petra Mohácsik 5, Csaba Fekete 6, Daofeng Li 7, Xiaoyun Xing 7, Ting Wang 7, Balázs Gereben 8, Antonio C Bianco 9
Affiliations
- PMID: 26508642
- PMCID: PMC4653175
- DOI: 10.1073/pnas.1508943112
Perinatal deiodinase 2 expression in hepatocytes defines epigenetic susceptibility to liver steatosis and obesity
Tatiana L Fonseca et al. Proc Natl Acad Sci U S A. 2015.
Abstract
Thyroid hormone binds to nuclear receptors and regulates gene transcription. Here we report that in mice, at around the first day of life, there is a transient surge in hepatocyte type 2 deiodinase (D2) that activates the prohormone thyroxine to the active hormone triiodothyronine, modifying the expression of ∼165 genes involved in broad aspects of hepatocyte function, including lipid metabolism. Hepatocyte-specific D2 inactivation (ALB-D2KO) is followed by a delay in neonatal expression of key lipid-related genes and a persistent reduction in peroxisome proliferator-activated receptor-γ expression. Notably, the absence of a neonatal D2 peak significantly modifies the baseline and long-term hepatic transcriptional response to a high-fat diet (HFD). Overall, changes in the expression of approximately 400 genes represent the HFD response in control animals toward the synthesis of fatty acids and triglycerides, whereas in ALB-D2KO animals, the response is limited to a very different set of only approximately 200 genes associated with reverse cholesterol transport and lipase activity. A whole genome methylation profile coupled to multiple analytical platforms indicate that 10-20% of these differences can be related to the presence of differentially methylated local regions mapped to sites of active/suppressed chromatin, thus qualifying as epigenetic modifications occurring as a result of neonatal D2 inactivation. The resulting phenotype of the adult ALB-D2KO mouse is dramatic, with greatly reduced susceptibility to diet-induced steatosis, hypertriglyceridemia, and obesity.
Keywords: deiodinase; lipids; obesity; steatosis; thyroid hormone.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Fig. 1.
Liver deiodinases in ALB-D2KO and control mice. (A) Relative Dio2 mRNA levels in embryonic (E12 and E15), neonatal (P1–P5) and adult livers. D2 mRNA levels are relative to 18S mRNA levels and normalized to E12. (B) Bright-field image of cresil violet- and eosin-counterstained P1 liver section. (C–H) Cresil violet-counterstained (C, E, and G) and dark-field (D, F, and H) images of D2 mRNA hybridization in mouse liver. (C and D) P1 mouse liver with D2-expressing cells distributed uniformly (higher-magnification view in C, Inset). (E and F) Background signal with sense probe in a P1 animal. (G and H) Adult hypothyroid mouse liver showing no signal after 6-wk exposure. (Scale bar: 50 μm.) (I) D2 activity in liver sonicates from neonatal ALB-D2KO and Global-D2KO mice (n = 5–7). (J) T3 production in intact hepatocytes of P1 and P5 animals (32). (K) Liver T3 content of P1 animals (32). All of the modifications are in supplanted material. Values are the mean ± SEM of three to seven independent samples. *P < 0.05, **P < 0.01 vs. controls.
Fig. 2.
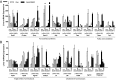
Liver expression of lipid-related genes at different ages in ALB-D2KO and control mice. mRNA data are expressed as in Fig. 1_A_. Genes are grouped as fatty acid biosynthesis and fatty acid oxidation (A) and triglyceride homeostasis and cholesterol homeostasis (B). Values are mean ± SEM of four to eight independent samples. Gene abbreviations are as indicated in SI Appendix, Tables S11 and S12. *P < 0.05, **P < 0.01 vs. same-age control animals.
Fig. 3.
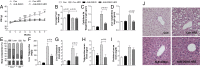
Metabolic parameters in ALB-D2KO mice maintained on an HFD. (A) Body weight evolution. (B) Weekly food intake. (C) Epididimal fat pad weight. (D) Liver weight. (E) Body composition as measured by DEXA at 48 h before admission to CLAMS. (F) Liver triglycerides. (G) Liver cholesterol. (H) Serum triglycerides. (I) Serum cholesterol. (J) Representative liver section stained with H&E. (Original magnification, 40×.) All entries are mean ± SEM of four to six animals. #P < 0.05, ###P < 0.001 vs. control; ***P < 0.001 vs. all groups.
Fig. 4.
Transcriptome and genome-wide DNA methylation profiling in adult ALB-D2KO mice. Venn diagram indicating overlapping between k4me1/3 + K36me3 (orange circle) or k27me3 (brown circle) containing histones and DMRs (green circle). Some of the gene sets contained in the overlapping areas (as identified by GREAT or MSigDB) are indicated. P values were calculated by a hypergeometric test in the R environment.
Fig. 5.
Gene expression analysis in liver of ALB-D2KO animals maintained on an HFD. Liver mRNA levels of candidate genes are shown. The results are relative to 18S mRNA levels and normalized to control chow diet animals. Values are mean ± SEM of five to eight independent samples. Gene abbreviations are provided in SI Appendix, Tables S11 and S12. #P < 0.05 vs. respective chow diet control; *P < 0.05 vs. respective HFD control.
References
- Andersen S, Bruun NH, Pedersen KM, Laurberg P. Biologic variation is important for interpretation of thyroid function tests. Thyroid. 2003;13(11):1069–1078. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- 2U01CA200060/CA/NCI NIH HHS/United States
- R01 HG007175/HG/NHGRI NIH HHS/United States
- 5R01ES024992/ES/NIEHS NIH HHS/United States
- U01 CA200060/CA/NCI NIH HHS/United States
- R01 ES024992/ES/NIEHS NIH HHS/United States
- 5R01HG007175/HG/NHGRI NIH HHS/United States
- R01 37021/PHS HHS/United States
- 5R01HG007354/HG/NHGRI NIH HHS/United States
- R25 DA027995/DA/NIDA NIH HHS/United States
- R01 65055/PHS HHS/United States
- R01 HG007354/HG/NHGRI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous