Gut mucosal microbiome across stages of colorectal carcinogenesis - PubMed (original) (raw)
Xiangchun Li 1 2, Haokui Zhou 3, Jianqiu Sheng 4, Sunny Hei Wong 1 2, William Ka Kai Wu 1 2 5, Siew Chien Ng 1 2, Ho Tsoi 1 2, Yujuan Dong 1 2, Ning Zhang 6, Yuqi He 4, Qian Kang 4, Lei Cao 1 2, Kunning Wang 1 2, Jingwan Zhang 1 2, Qiaoyi Liang 1 2, Jun Yu 1 2, Joseph J Y Sung 1 2
Affiliations
- PMID: 26515465
- PMCID: PMC4640069
- DOI: 10.1038/ncomms9727
Gut mucosal microbiome across stages of colorectal carcinogenesis
Geicho Nakatsu et al. Nat Commun. 2015.
Abstract
Gut microbial dysbiosis contributes to the development of colorectal cancer (CRC). Here we catalogue the microbial communities in human gut mucosae at different stages of colorectal tumorigenesis. We analyse the gut mucosal microbiome of 47 paired samples of adenoma and adenoma-adjacent mucosae, 52 paired samples of carcinoma and carcinoma-adjacent mucosae and 61 healthy controls. Probabilistic partitioning of relative abundance profiles reveals that a metacommunity predominated by members of the oral microbiome is primarily associated with CRC. Analysis of paired samples shows differences in community configurations between lesions and the adjacent mucosae. Correlations of bacterial taxa indicate early signs of dysbiosis in adenoma, and co-exclusive relationships are subsequently more common in cancer. We validate these alterations in CRC-associated microbiome by comparison with two previously published data sets. Our results suggest that a taxonomically defined microbial consortium is implicated in the development of CRC.
Figures
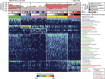
Figure 1. Characterization of 16S rRNA gene catalogue for mucosal microbial communities in colorectal carcinogenesis.
Fitting microbiome data to DMM models defined five metacommunities. Reads that are considered as being potentially originated from oral strains or known pathogenic strains in the human gut were classified against the 16S rRNA gene collections from the Human Oral Microbiome (HOM; version 13) database and PATRIC bacterial pathogen database as defined by pseudo-bootstrapped (_n_=1,000) confidence scores of 100 at species-level taxa or deeper, using the naive Bayesian classifier. The panels of metacommunity markers are ranked in the descending order of linear discriminant analysis scores from top to bottom. Columns represent microbiome profiles (arcsine square root-transformed) of 269 mucosal biopsies from individuals with or without adenoma or adenocarcinomas. (MCPI<0 for changes characteristic of adenomas; MCPI>0 for changes characteristic of carcinomas).
Figure 2. Validations of metacommunity markers in independent cohorts.
(a,b) Fold-change analyses in paired carcinoma and carcinoma-adjacent samples in two additional cohorts demonstrated significant agreement with our discovery cohort: (a) Kostic et al. data set (_n_=74) and (b) Zeller et al. data set (_n_=48). Shown are adjusted _R_2 and P values for goodness of fit from multiple linear regression models. (c) Real-time PCR amplifications of the most abundant sequences of representative bacterial phylotypes showed consistent enrichments in an additional Chinese cohort consisting of 207 mucosal biopsies (normal control, _n_=25; adenoma, _n_=41; adenocarcinoma, _n_=50). Error bars represent s.e.m. P values from Mann–Whitney _U_-tests are adjusted by Benjamini-Hochberg (BH) step-up procedure; *q<0.05; **q<0.01; ***q<0.001; ****q<0.0001.
Figure 3. Community-wide alterations of microbiome profiles are important aspects of multistage colorectal tumour progression.
(a) Discordance of taxonomic configurations between lesions and lesion-adjacent tissues was significantly associated with the metacommunities identified within carcinoma. Shown are mean P values from 1,000 iterations of Fisher's exact tests with Monte Carlo simulation (10,000 replicates). (b) Percentages of change between metacommunities from lesion-adjacent mucosae to lesions within each clinicopathologic stage of tumours. LGDP, colorectal polyps with low-grade dysplasia (_n_=39); HGDP, colorectal polyps with high-grade dysplasia (_n_=13); ECRC, early-stage CRC (_n_=26); LCRC, late-stage CRC (_n_=26). (c) Significances of fold change in metacommunity markers, as estimated by paired Mann–Whitney _U_-tests, were greatest at early-stage CRC.
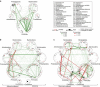
Figure 4. Microbial community ecology at mucosal interface are different across stages of colorectal carcinogenesis.
(a–c) Correlation network of taxonomic partners in: (a) normal (_n_=61), (b) adenomatous polyps (_n_=52) and (c) cancerous mucosae (_n_=52). Correlation coefficients were estimated and corrected for compositional effects using the SparCC algorithm. A subset of correlations with strengths of at least 0.3 was selected for visualization. Node size represents mean taxon abundance in each mucosal phenotype; metacommunity markers are denoted by node numbers accordingly. Taxa that are classified as members of the same bacterial phylum are encircled by dashed lines.
Similar articles
- Altered intestinal microbiota associated with colorectal cancer.
Zhang H, Chang Y, Zheng Q, Zhang R, Hu C, Jia W. Zhang H, et al. Front Med. 2019 Aug;13(4):461-470. doi: 10.1007/s11684-019-0695-7. Epub 2019 Jun 28. Front Med. 2019. PMID: 31250341 - Fecal Metabolomic Signatures in Colorectal Adenoma Patients Are Associated with Gut Microbiota and Early Events of Colorectal Cancer Pathogenesis.
Kim M, Vogtmann E, Ahlquist DA, Devens ME, Kisiel JB, Taylor WR, White BA, Hale VL, Sung J, Chia N, Sinha R, Chen J. Kim M, et al. mBio. 2020 Feb 18;11(1):e03186-19. doi: 10.1128/mBio.03186-19. mBio. 2020. PMID: 32071266 Free PMC article. - Changes in gut microbiota and plasma inflammatory factors across the stages of colorectal tumorigenesis: a case-control study.
Zhang Y, Yu X, Yu E, Wang N, Cai Q, Shuai Q, Yan F, Jiang L, Wang H, Liu J, Chen Y, Li Z, Jiang Q. Zhang Y, et al. BMC Microbiol. 2018 Aug 29;18(1):92. doi: 10.1186/s12866-018-1232-6. BMC Microbiol. 2018. PMID: 30157754 Free PMC article. - Microbiome in Colorectal Cancer: How to Get from Meta-omics to Mechanism?
Ternes D, Karta J, Tsenkova M, Wilmes P, Haan S, Letellier E. Ternes D, et al. Trends Microbiol. 2020 May;28(5):401-423. doi: 10.1016/j.tim.2020.01.001. Epub 2020 Feb 13. Trends Microbiol. 2020. PMID: 32298617 Review. - Gut mycobiome: A promising target for colorectal cancer.
Qin X, Gu Y, Liu T, Wang C, Zhong W, Wang B, Cao H. Qin X, et al. Biochim Biophys Acta Rev Cancer. 2021 Jan;1875(1):188489. doi: 10.1016/j.bbcan.2020.188489. Epub 2020 Dec 3. Biochim Biophys Acta Rev Cancer. 2021. PMID: 33278512 Review.
Cited by
- Bacteria-related changes in host DNA methylation and the risk for CRC.
Sobhani I, Rotkopf H, Khazaie K. Sobhani I, et al. Gut Microbes. 2020 Nov 9;12(1):1800898. doi: 10.1080/19490976.2020.1800898. Gut Microbes. 2020. PMID: 32931352 Free PMC article. Review. - Could environmental exposure and climate change Be a key factor in the rising incidence of early onset colorectal cancer?
AlZaabi A, Younus HA, Al-Reasi HA, Al-Hajri R. AlZaabi A, et al. Heliyon. 2024 Aug 10;10(16):e35935. doi: 10.1016/j.heliyon.2024.e35935. eCollection 2024 Aug 30. Heliyon. 2024. PMID: 39258208 Free PMC article. Review. - The Macrophages-Microbiota Interplay in Colorectal Cancer (CRC)-Related Inflammation: Prognostic and Therapeutic Significance.
Mola S, Pandolfo C, Sica A, Porta C. Mola S, et al. Int J Mol Sci. 2020 Sep 18;21(18):6866. doi: 10.3390/ijms21186866. Int J Mol Sci. 2020. PMID: 32962159 Free PMC article. Review. - Clinical Implications of the Associations Between Intestinal Microbiome and Colorectal Cancer Progression.
Chen Y, Yang Y, Gu J. Chen Y, et al. Cancer Manag Res. 2020 Jun 9;12:4117-4128. doi: 10.2147/CMAR.S240108. eCollection 2020. Cancer Manag Res. 2020. PMID: 32606919 Free PMC article. Review. - Understanding the role of the gut microbiome in gastrointestinal cancer: A review.
Ağagündüz D, Cocozza E, Cemali Ö, Bayazıt AD, Nanì MF, Cerqua I, Morgillo F, Saygılı SK, Berni Canani R, Amero P, Capasso R. Ağagündüz D, et al. Front Pharmacol. 2023 Jan 24;14:1130562. doi: 10.3389/fphar.2023.1130562. eCollection 2023. Front Pharmacol. 2023. PMID: 36762108 Free PMC article. Review.
References
- Irrazabal T., Belcheva A., Girardin S. E., Martin A. & Philpott D. J. The multifaceted role of the intestinal microbiota in colon cancer. Mol. Cell 54, 309–320 (2014). - PubMed
- Belcheva A. et al. Gut microbial metabolism drives transformation of Msh2-deficient colon epithelial cells. Cell 158, 288–299 (2014). - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical