Transcription factor trapping by RNA in gene regulatory elements - PubMed (original) (raw)
Transcription factor trapping by RNA in gene regulatory elements
Alla A Sigova et al. Science. 2015.
Abstract
Transcription factors (TFs) bind specific sequences in promoter-proximal and -distal DNA elements to regulate gene transcription. RNA is transcribed from both of these DNA elements, and some DNA binding TFs bind RNA. Hence, RNA transcribed from regulatory elements may contribute to stable TF occupancy at these sites. We show that the ubiquitously expressed TF Yin-Yang 1 (YY1) binds to both gene regulatory elements and their associated RNA species across the entire genome. Reduced transcription of regulatory elements diminishes YY1 occupancy, whereas artificial tethering of RNA enhances YY1 occupancy at these elements. We propose that RNA makes a modest but important contribution to the maintenance of certain TFs at gene regulatory elements and suggest that transcription of regulatory elements produces a positive-feedback loop that contributes to the stability of gene expression programs.
Copyright © 2015, American Association for the Advancement of Science.
Figures
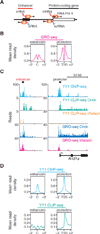
Fig. 1
YY1 binds to DNA and RNA at transcriptional regulatory elements. (A) Cartoon depicting divergent transcription at enhancers and promoters in mammalian cells. (B) Alignment of GRO-seq reads at all enhancers and promoters in ESCs. Enhancers were defined as in (23). The x-axis indicates distance from either the enhancer center (C) or the transcription start site (TSS) in kilobases. The y-axis indicates average density of uniquely mapped GRO-seq reads per genomic bin. (C) Gene tracks for the Arid1a gene and enhancer showing ChIP-seq and CLIP-seq data for bio-YY1 cells, as well as GRO-seq reads for mESCs. (D) Mean read density of YY1 ChIP-seq and CLIP-seq reads at enhancers and promoters of all RefSeq genes in ESCs.
Fig. 2
YY1 binds to DNA and RNA in vitro. (A) Left panel: EMSA of YY1-DNA complexes at different concentrations of recombinant YY1. 5 nM of radioactively labelled 30-bp DNA probe derived from the promoter region of Arid1a gene containing a consensus YY1 binding motif (CTCTTCTCTCTTAAAATGGCTGCCTGTCTG) was incubated with increasing concentrations of recombinant murine YY1 protein. Right panel: EMSA of YY1-RNA complexes at different concentrations of recombinant YY1. 5 nM of radioactively labeled 30-nt RNA probe derived from the same region of the Arid1a gene was incubated with increasing concentrations of recombinant YY1 protein. (B) Graph depicting relationship between the fraction of radioactively labeled DNA or RNA probe bound and the concentration of recombinant YY1 in the binding reaction.
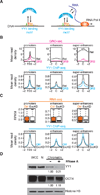
Fig. 3
Perturbation of RNA levels affects YY1 binding to DNA. (A) Cartoon depicting hypothesis that RNA transcribed from regulatory elements enhances occupancy of these elements by TFs capable of binding both DNA and RNA. (B) (Top) GRO-seq reads (24) at promoters, enhancers, and super-enhancer constituents in cells before (DRB) and after release (Rel) from transcriptional inhibition by DRB. (Bottom) YY1 ChIP-seq reads at promoters, enhancers and super-enhancer constituents in cells before (DRB) and after release (Rel) from transcriptional inhibition by DRB. Increase in YY1 binding after release from DRB inhibition is significant: _p_-value < 3.6x10−207 for promoters, _p_-value < 1.6x10−214 for enhancers, _p_-value < 9.8x10−37 for super-enhancers. (C) (Top) Box plots depicting RNA-seq data for ribo-depleted total RNA at promoters, enhancers, and super-enhancers in ESCs after targeting with control (Ctrl) or Exosc3 (ExoKD) shRNA. (Bottom) Alignment of YY1 ChIP-seq reads at promoters, enhancers and super-enhancers in ESCs after targeting with control (Ctrl) or Exosc3 (ExoKD) shRNA. The decrease in YY1 binding in ExoKD ESCs is significant: _p_-value < 8.1x10−9 for promoters _p_-value < 1.8x10−27 for enhancers, _p_-value < 3.3x10−5 for super-enhancers. (D) Western blot analysis of YY1, OCT4, and histone H3 levels in whole-cell extracts (WCE), nuclei (N), and a nuclear chromatin preparation before and after RNase A treatment. Histone H3 serves as a loading control and OCT4 serves as a negative control. Quantitation of the relative levels of YY1 and OCT4 are noted.
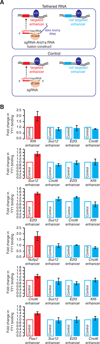
Fig. 4
Tethering of RNA adjacent an YY1 DNA binding site enhances binding of YY1 to the genome in vivo. (A) Strategy for tethering of RNA in the vicinity of an YY1 binding site at enhancers in vivo. (B) ChIP-qPCR analysis of YY1 binding at six targeted (red) and three not targeted (blue) enhancers in three independent experiments. The y-axis indicates fold change in YY1 binding in ESCs expressing the sgRNA-Arid1a RNA fusion construct relative to cells expressing the control sgRNA targeted to the same locus. The difference in YY1 binding was significant for the targeted enhancers: Klf5 (_p_-value=0.03), Suz12 (_p_-value=0.01), E2f3 (_p_-value=0.01), Nufip2 (_p_-value=0.03), Cnot6 (_p_-value=0.03), and Pias1 (_p_-value=0.01), but not for the not targeted enhancers.
Similar articles
- YY1 Is a Structural Regulator of Enhancer-Promoter Loops.
Weintraub AS, Li CH, Zamudio AV, Sigova AA, Hannett NM, Day DS, Abraham BJ, Cohen MA, Nabet B, Buckley DL, Guo YE, Hnisz D, Jaenisch R, Bradner JE, Gray NS, Young RA. Weintraub AS, et al. Cell. 2017 Dec 14;171(7):1573-1588.e28. doi: 10.1016/j.cell.2017.11.008. Epub 2017 Dec 7. Cell. 2017. PMID: 29224777 Free PMC article. - Binding of YY1 to the proximal region of the murine beta interferon promoter is essential to allow CBP recruitment and K8H4/K14H3 acetylation on the promoter region after virus infection.
Mokrani H, Sharaf el Dein O, Mansuroglu Z, Bonnefoy E. Mokrani H, et al. Mol Cell Biol. 2006 Nov;26(22):8551-61. doi: 10.1128/MCB.00420-06. Epub 2006 Sep 5. Mol Cell Biol. 2006. PMID: 16954376 Free PMC article. - Role of non-coding RNA transcription around gene regulatory elements in transcription factor recruitment.
Takemata N, Ohta K. Takemata N, et al. RNA Biol. 2017 Jan 2;14(1):1-5. doi: 10.1080/15476286.2016.1248020. Epub 2016 Oct 20. RNA Biol. 2017. PMID: 27763805 Free PMC article. Review. - In pursuit of design principles of regulatory sequences.
Levo M, Segal E. Levo M, et al. Nat Rev Genet. 2014 Jul;15(7):453-68. doi: 10.1038/nrg3684. Epub 2014 Jun 10. Nat Rev Genet. 2014. PMID: 24913666 Review.
Cited by
- Re-analysis of CLAP data affirms PRC2 as an RNA binding protein.
Lee Y, Blum R, Rosenberg M, Lee JT. Lee Y, et al. bioRxiv [Preprint]. 2024 Sep 20:2024.09.19.613009. doi: 10.1101/2024.09.19.613009. bioRxiv. 2024. PMID: 39345380 Free PMC article. Preprint. - RNA Interactions Are Essential for CTCF-Mediated Genome Organization.
Saldaña-Meyer R, Rodriguez-Hernaez J, Escobar T, Nishana M, Jácome-López K, Nora EP, Bruneau BG, Tsirigos A, Furlan-Magaril M, Skok J, Reinberg D. Saldaña-Meyer R, et al. Mol Cell. 2019 Nov 7;76(3):412-422.e5. doi: 10.1016/j.molcel.2019.08.015. Epub 2019 Sep 12. Mol Cell. 2019. PMID: 31522988 Free PMC article. - Analysis of estrogen-regulated enhancer RNAs identifies a functional motif required for enhancer assembly and gene expression.
Hou TY, Kraus WL. Hou TY, et al. Cell Rep. 2022 Jun 14;39(11):110944. doi: 10.1016/j.celrep.2022.110944. Cell Rep. 2022. PMID: 35705040 Free PMC article. - Deciphering shared attributes of plant long non-coding RNAs through a comparative computational approach.
Yadav VK, Jalmi SK, Tiwari S, Kerkar S. Yadav VK, et al. Sci Rep. 2023 Sep 12;13(1):15101. doi: 10.1038/s41598-023-42420-7. Sci Rep. 2023. PMID: 37699996 Free PMC article. - Genome-wide analysis of the interplay between chromatin-associated RNA and 3D genome organization in human cells.
Calandrelli R, Wen X, Charles Richard JL, Luo Z, Nguyen TC, Chen CJ, Qi Z, Xue S, Chen W, Yan Z, Wu W, Zaleta-Rivera K, Hu R, Yu M, Wang Y, Li W, Ma J, Ren B, Zhong S. Calandrelli R, et al. Nat Commun. 2023 Oct 16;14(1):6519. doi: 10.1038/s41467-023-42274-7. Nat Commun. 2023. PMID: 37845234 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous