A Novel Saccharomyces cerevisiae FG Nucleoporin Mutant Collection for Use in Nuclear Pore Complex Functional Experiments - PubMed (original) (raw)
A Novel Saccharomyces cerevisiae FG Nucleoporin Mutant Collection for Use in Nuclear Pore Complex Functional Experiments
Rebecca L Adams et al. G3 (Bethesda). 2015.
Abstract
FG nucleoporins (Nups) are the class of proteins that both generate the permeability barrier and mediate selective transport through the nuclear pore complex (NPC). The FG Nup family has 11 members in Saccharomyces cerevisiae, and the study of mutants lacking different FG domains has been instrumental in testing transport models. To continue analyzing the distinct functional roles of FG Nups in vivo, additional robust genetic tools are required. Here, we describe a novel collection of S. cerevisiae mutant strains in which the FG domains of different groups of Nups are absent (Δ) in the greatest number documented to date. Using this plasmid-based ΔFG strategy, we find that a GLFG domain-only pore is sufficient for viability. The resulting extensive plasmid and strain resources are available to the scientific community for future in-depth in vivo studies of NPC transport.
Keywords: FG nucleoporin; S. cerevisiae; nuclear pore complex.
Copyright © 2016 Adams et al.
Figures
Figure 1
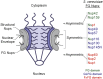
Schematic of NPC depicting relative structural location of FG Nups, based on Rout et al. (2000) with the image adapted from Adams and Wente (2013). FG Nups are color-coded based on the type of FG repeats enriched in their FG domains: Green, FG; Blue, GLFG; Red, FxFG. Nsp1 contains both FG and FxFG domains, and Nup116 contains both FG and GLFG domains.
Figure 2
(A) Schematic of Δ_FG_ plasmid construction. Centromeric plasmids encoding a WT NUP gene with its endogenous 5′ and 3′ UTR were PCR amplified with primers that annealed outside of the FG domain and generated a unique in-frame restriction site. PCR products were cut and ligated back together to generate the Δ_FG_ plasmid. Δ_FG nups_ or WT NUPS were subcloned into one plasmid encoding multiple genes (Table 2). (B) Schematic depicting Δ_FG_ strain construction. Plasmids harboring multiple NUP genes were transformed into parent strains followed by disruption of the chromosomal ORF with sequence encoding floxed Schizosaccharomyces pombe HIS5 (SpHIS5). SpHIS5 was then looped out by transformation with a plasmid for inducible expression of Cre recombinase. Iterative transformation, disruption, and SpHIS5 recycling cycles were used to generate indicated strains. Strains were subsequently transformed with Δ_FG nup_ plasmids and counterselected.
Figure 3
Construction history of Δ_FG_ deletion strains. Beginning with a WT strain, NSP1, NUP49, and NUP57 were individually deleted in the presence of a WT NUP vector. Strains were mated and sporulated to generate a triple null, and pSW3643 was transformed with counterselection of single gene-encoding plasmids to generate SWY4684. SWY4684 was transformed with pSW3547, and pSW3643 was counterselected on with the TRP1 counterselective drug 5-FAA to generate SWY4683. SWY4684 was transformed with pSW3641, and NUP1, NUP2, and NUP60 were deleted iteratively to generate SWY4688. SWY4688 was transformed with pSW3547, and pSW3643 was counterselected on 5-FAA to generate SWY4690. LYS2 was deleted from SWY4688 with a floxed SpHIS5 cassette, which was recombined. This strain was then transformed with pSW3646, and NUP42 and NUP159 were deleted iteratively to generate SWY4779. SWY4779 was transformed with pSW3547, and pSW3643 was counterselected on 5-FAA to generate SWY6359. SWY4779 was transformed with pSW3642, and colonies with spontaneous loss of LEU2 were selected to generate SWY6360. SWY6360 was transformed with pSW3547, and pSW3643 was counterselected on 5-FAA to generate SWY6361. Additional strain and plasmid information is described in Table 1 and Table 2.
Figure 4
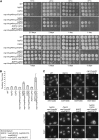
(A) Growth analysis of Δ_FG_ strains at different temperatures. Yeast strains were grown at 23° to midlog phase and five-fold serially diluted on YPD plates for growth at the indicated temperature for 1–7 days. (B) Liquid growth analysis of Δ_FG_ strains. Yeast strains were grown at 23° to early log phase, and OD600 was analyzed to determine doubling times. Error bars indicate standard deviation from three replicates. (C) Nup116 is properly assembled into NPCs of Δ_FG_ strains. Indicated strains were grown at 23° to midlog phase and processed for indirect immunofluorescence microscopy using the anti-(α)-Nup116-CTD antibodies. DAPI staining marks the nucleus. N_Δ_FG C_Δ_FG nsp1/nup49/nup57_Δ_FG was scaled independently due to increased cellular autofluorescence. Scale bar, 5 μm.
Similar articles
- Nucleoporin's Like Charge Regions Are Major Regulators of FG Coverage and Dynamics Inside the Nuclear Pore Complex.
Peyro M, Soheilypour M, Ghavami A, Mofrad MR. Peyro M, et al. PLoS One. 2015 Dec 11;10(12):e0143745. doi: 10.1371/journal.pone.0143745. eCollection 2015. PLoS One. 2015. PMID: 26658558 Free PMC article. - Natively unfolded nucleoporins gate protein diffusion across the nuclear pore complex.
Patel SS, Belmont BJ, Sante JM, Rexach MF. Patel SS, et al. Cell. 2007 Apr 6;129(1):83-96. doi: 10.1016/j.cell.2007.01.044. Cell. 2007. PMID: 17418788 - The FG-repeat asymmetry of the nuclear pore complex is dispensable for bulk nucleocytoplasmic transport in vivo.
Zeitler B, Weis K. Zeitler B, et al. J Cell Biol. 2004 Nov 22;167(4):583-90. doi: 10.1083/jcb.200407156. J Cell Biol. 2004. PMID: 15557115 Free PMC article. - Biomechanics of the transport barrier in the nuclear pore complex.
Stanley GJ, Fassati A, Hoogenboom BW. Stanley GJ, et al. Semin Cell Dev Biol. 2017 Aug;68:42-51. doi: 10.1016/j.semcdb.2017.05.007. Epub 2017 May 12. Semin Cell Dev Biol. 2017. PMID: 28506890 Review. - Flexible gates: dynamic topologies and functions for FG nucleoporins in nucleocytoplasmic transport.
Terry LJ, Wente SR. Terry LJ, et al. Eukaryot Cell. 2009 Dec;8(12):1814-27. doi: 10.1128/EC.00225-09. Epub 2009 Oct 2. Eukaryot Cell. 2009. PMID: 19801417 Free PMC article. Review.
Cited by
- Simple rules for passive diffusion through the nuclear pore complex.
Timney BL, Raveh B, Mironska R, Trivedi JM, Kim SJ, Russel D, Wente SR, Sali A, Rout MP. Timney BL, et al. J Cell Biol. 2016 Oct 10;215(1):57-76. doi: 10.1083/jcb.201601004. Epub 2016 Oct 3. J Cell Biol. 2016. PMID: 27697925 Free PMC article. - The yeast Ty1 retrotransposon requires components of the nuclear pore complex for transcription and genomic integration.
Manhas S, Ma L, Measday V. Manhas S, et al. Nucleic Acids Res. 2018 Apr 20;46(7):3552-3578. doi: 10.1093/nar/gky109. Nucleic Acids Res. 2018. PMID: 29514267 Free PMC article. - Diameter dependence of transport through nuclear pore complex mimics studied using optical nanopores.
Klughammer N, Barth A, Dekker M, Fragasso A, Onck PR, Dekker C. Klughammer N, et al. Elife. 2024 Feb 20;12:RP87174. doi: 10.7554/eLife.87174. Elife. 2024. PMID: 38376900 Free PMC article. - Natively Unfolded FG Repeats Stabilize the Structure of the Nuclear Pore Complex.
Onischenko E, Tang JH, Andersen KR, Knockenhauer KE, Vallotton P, Derrer CP, Kralt A, Mugler CF, Chan LY, Schwartz TU, Weis K. Onischenko E, et al. Cell. 2017 Nov 2;171(4):904-917.e19. doi: 10.1016/j.cell.2017.09.033. Epub 2017 Oct 12. Cell. 2017. PMID: 29033133 Free PMC article. - An introduction to dynamic nucleoporins in Leishmania species: Novel targets for tropical-therapeutics.
Dubey AK, Kumar P, Mandal D, Ravichandiran V, Singh SK. Dubey AK, et al. J Parasit Dis. 2022 Dec;46(4):1176-1191. doi: 10.1007/s12639-022-01515-0. Epub 2022 Jul 25. J Parasit Dis. 2022. PMID: 36457769 Free PMC article. Review.
References
- Alber F., Dokudovskaya S., Veenhoff L. M., Zhang W., Kipper J., et al. , 2007. The molecular architecture of the nuclear pore complex. Nature 450: 695–701. - PubMed
- Chug H., Trakhanov S., Hulsmann B. B., Pleiner T., Gorlich D., 2015. Crystal structure of the metazoan Nup62•Nup58•Nup54 nucleoporin complex. Science. 350: 106–110 - PubMed
- Davis L. I., Fink G. R., 1990. The NUP1 gene encodes an essential component of the yeast nuclear pore complex. Cell 61: 965–978. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R37 GM051219/GM/NIGMS NIH HHS/United States
- T32 HD007502/HD/NICHD NIH HHS/United States
- R37GM051219/GM/NIGMS NIH HHS/United States
- T32HD007502/HD/NICHD NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases