CellProfiler Tracer: exploring and validating high-throughput, time-lapse microscopy image data - PubMed (original) (raw)
CellProfiler Tracer: exploring and validating high-throughput, time-lapse microscopy image data
Mark-Anthony Bray et al. BMC Bioinformatics. 2015.
Abstract
Background: Time-lapse analysis of cellular images is an important and growing need in biology. Algorithms for cell tracking are widely available; what researchers have been missing is a single open-source software package to visualize standard tracking output (from software like CellProfiler) in a way that allows convenient assessment of track quality, especially for researchers tuning tracking parameters for high-content time-lapse experiments. This makes quality assessment and algorithm adjustment a substantial challenge, particularly when dealing with hundreds of time-lapse movies collected in a high-throughput manner.
Results: We present CellProfiler Tracer, a free and open-source tool that complements the object tracking functionality of the CellProfiler biological image analysis package. Tracer allows multi-parametric morphological data to be visualized on object tracks, providing visualizations that have already been validated within the scientific community for time-lapse experiments, and combining them with simple graph-based measures for highlighting possible tracking artifacts.
Conclusions: CellProfiler Tracer is a useful, free tool for inspection and quality control of object tracking data, available from http://www.cellprofiler.org/tracer/.
Figures
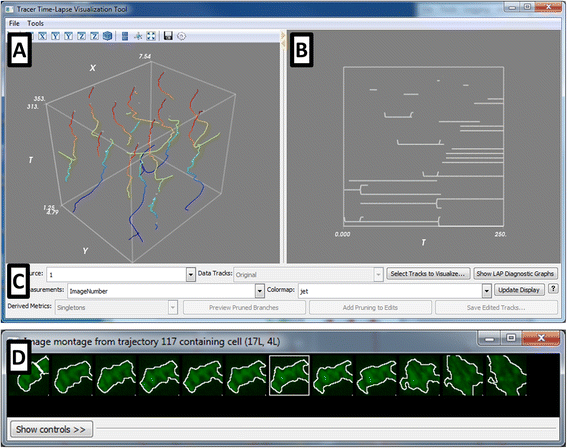
Fig. 1
The CellProfiler Tracer interface. The user interface is divided into the (a) XYT panel, showing the object trajectories in (x,y,t) coordinates, color-coded here by the frame number; the trajectories can be color-coded to be any cell measurement of interest; (b) the lineage tree panel, highlighting the ancestor/progeny relationships corresponding to the trajectories in (a), and (c) the control panel containing various display tools. Other visualizations include (d) synchrograms of selected cells, as well as heatmaps (shown in Fig. 2)
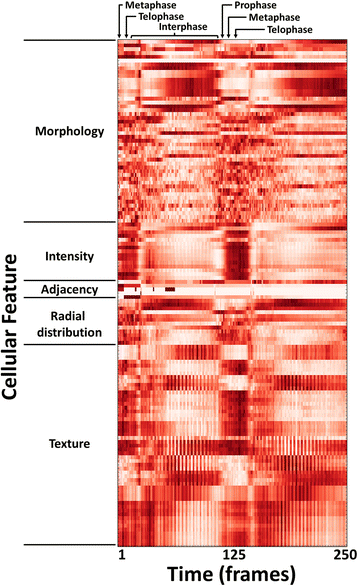
Fig. 2
Heatmap of high-content cellular time-lapse measurements. The per-nucleus measurements from a Drosophila time-lapse movie are averaged over all nuclei for each timepoint; the measurements were collected by CellProfiler software. Feature values were normalized from 0 to 1 for visualization purposes. Feature names were omitted for conciseness but are provided in the Tracer display; the features shown are listed in order in the Additional file 3: Table S1, and are further described in the CellProfiler documentation
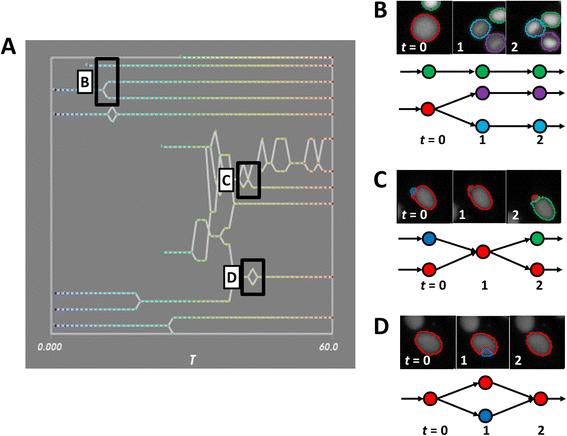
Fig. 3
Schematics of tracking errors. a An inset of the lineage panel for a movie of MCF-7 cells, with various tracking topologies highlighted. b-d Tracking errors are reflected in synchrograms of MCF-7 nuclei (top panel) and graph topologies (bottom panel) with color indicating the unique object label. b Typical graphs with no tracking errors. c Mis-segmentation of neighboring objects produces transient merging and erroneous object creation. d A brief mis-segmentation of an object results in a transient (and incorrect) split
Similar articles
- Traxtile: Interactive editing of cell tracks in time-lapse images.
Braun BS. Braun BS. Biotechniques. 2015 Aug 1;59(2):82-6. doi: 10.2144/000114318. eCollection 2015 Aug. Biotechniques. 2015. PMID: 26260086 Free PMC article. - A robust algorithm for segmenting and tracking clustered cells in time-lapse fluorescent microscopy.
Tarnawski W, Kurtcuoglu V, Lorek P, Bodych M, Rotter J, Muszkieta M, Piwowar Ł, Poulikakos D, Majkowski M, Ferrari A. Tarnawski W, et al. IEEE J Biomed Health Inform. 2013 Jul;17(4):862-9. doi: 10.1109/JBHI.2013.2262233. IEEE J Biomed Health Inform. 2013. PMID: 25055315 - STrack: A Tool to Simply Track Bacterial Cells in Microscopy Time-Lapse Images.
Todorov H, Miguel Trabajo T, van der Meer JR. Todorov H, et al. mSphere. 2023 Apr 20;8(2):e0065822. doi: 10.1128/msphere.00658-22. Epub 2023 Mar 20. mSphere. 2023. PMID: 36939355 Free PMC article. - Untangling cell tracks: Quantifying cell migration by time lapse image data analysis.
Svensson CM, Medyukhina A, Belyaev I, Al-Zaben N, Figge MT. Svensson CM, et al. Cytometry A. 2018 Mar;93(3):357-370. doi: 10.1002/cyto.a.23249. Epub 2017 Oct 4. Cytometry A. 2018. PMID: 28976646 Review. - Methods for cell and particle tracking.
Meijering E, Dzyubachyk O, Smal I. Meijering E, et al. Methods Enzymol. 2012;504:183-200. doi: 10.1016/B978-0-12-391857-4.00009-4. Methods Enzymol. 2012. PMID: 22264535 Review.
Cited by
- Calcium-vesicles perform active diffusion in the sea urchin embryo during larval biomineralization.
Winter MR, Morgulis M, Gildor T, Cohen AR, Ben-Tabou de-Leon S. Winter MR, et al. PLoS Comput Biol. 2021 Feb 22;17(2):e1008780. doi: 10.1371/journal.pcbi.1008780. eCollection 2021 Feb. PLoS Comput Biol. 2021. PMID: 33617532 Free PMC article. - Community standards for open cell migration data.
Gonzalez-Beltran AN, Masuzzo P, Ampe C, Bakker GJ, Besson S, Eibl RH, Friedl P, Gunzer M, Kittisopikul M, Dévédec SEL, Leo S, Moore J, Paran Y, Prilusky J, Rocca-Serra P, Roudot P, Schuster M, Sergeant G, Strömblad S, Swedlow JR, van Erp M, Van Troys M, Zaritsky A, Sansone SA, Martens L. Gonzalez-Beltran AN, et al. Gigascience. 2020 May 1;9(5):giaa041. doi: 10.1093/gigascience/giaa041. Gigascience. 2020. PMID: 32396199 Free PMC article. - Inferring cell state by quantitative motility analysis reveals a dynamic state system and broken detailed balance.
Kimmel JC, Chang AY, Brack AS, Marshall WF. Kimmel JC, et al. PLoS Comput Biol. 2018 Jan 16;14(1):e1005927. doi: 10.1371/journal.pcbi.1005927. eCollection 2018 Jan. PLoS Comput Biol. 2018. PMID: 29338005 Free PMC article. - Antibody surface mobility amplifies FcγR signaling via Arp2/3 during phagocytosis.
Jo S, Fischer BR, Cronin NM, Nurmalasari NPD, Loyd YM, Kerkvliet JG, Bailey EM, Anderson RB, Scott BL, Hoppe AD. Jo S, et al. Biophys J. 2024 Aug 6;123(15):2312-2327. doi: 10.1016/j.bpj.2024.01.036. Epub 2024 Feb 5. Biophys J. 2024. PMID: 38321740 - Emerging machine learning approaches to phenotyping cellular motility and morphodynamics.
Choi HJ, Wang C, Pan X, Jang J, Cao M, Brazzo JA 3rd, Bae Y, Lee K. Choi HJ, et al. Phys Biol. 2021 Jun 17;18(4):10.1088/1478-3975/abffbe. doi: 10.1088/1478-3975/abffbe. Phys Biol. 2021. PMID: 33971636 Free PMC article. Review.
References
- Neumann B, Walter T, Hériché J-K, Bulkescher J, Erfle H, Conrad C, Rogers P, Poser I, Held M, Liebel U, Cetin C, Sieckmann F, Pau G, Kabbe R, Wünsche A, Satagopam V, Schmitz MHA, Chapuis C, Gerlich DW, Schneider R, Eils R, Huber W, Peters J-M, Hyman AA, Durbin R, Pepperkok R, Ellenberg J. Phenotypic profiling of the human genome by time-lapse microscopy reveals cell division genes. Nature. 2010;464:721–727. doi: 10.1038/nature08869. - DOI - PMC - PubMed
- Carpenter AE, Jones TR, Lamprecht MR, Clarke C, Kang IH, Friman O, Guertin DA, Chang JH, Lindquist RA, Moffat J, Golland P, Sabatini DM. Cell Profiler: image analysis software for identifying and quantifying cell phenotypes. Genome Biol. 2006;7:R100. doi: 10.1186/gb-2006-7-10-r100. - DOI - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources