Endothelial Cell Proteomic Response to Rickettsia conorii Infection Reveals Activation of the Janus Kinase (JAK)-Signal Transducer and Activator of Transcription (STAT)-Inferferon Stimulated Gene (ISG)15 Pathway and Reprogramming Plasma Membrane Integrin/Cadherin Signaling - PubMed (original) (raw)
Endothelial Cell Proteomic Response to Rickettsia conorii Infection Reveals Activation of the Janus Kinase (JAK)-Signal Transducer and Activator of Transcription (STAT)-Inferferon Stimulated Gene (ISG)15 Pathway and Reprogramming Plasma Membrane Integrin/Cadherin Signaling
Yingxin Zhao et al. Mol Cell Proteomics. 2016 Jan.
Abstract
Rickettsia conorii is the etiologic agent of Mediterranean spotted fever, a re-emerging infectious disease with significant mortality. This Gram-negative, obligately intracellular pathogen is transmitted via tick bites, resulting in disseminated vascular endothelial cell infection with vascular leakage. In the infected human, Rickettsia conorii infects endothelial cells, stimulating expression of cytokines and pro-coagulant factors. However, the integrated proteomic response of human endothelial cells to R. conorii infection is not known. In this study, we performed quantitative proteomic profiling of primary human umbilical vein endothelial cells (HUVECs) with established R conorii infection versus those stimulated with endotoxin (LPS) alone. We observed differential expression of 55 proteins in HUVEC whole cell lysates. Of these, we observed induction of signal transducer and activator of transcription (STAT)1, MX dynamin-like GTPase (MX1), and ISG15 ubiquitin-like modifier, indicating activation of the JAK-STAT signaling pathway occurs in R. conorii-infected HUVECs. The down-regulated proteins included those involved in the pyrimidine and arginine biosynthetic pathways. A highly specific biotinylated cross-linking enrichment protocol was performed to identify dysregulation of 11 integral plasma membrane proteins that included up-regulated expression of a sodium/potassium transporter and down-regulation of α-actin 1. Analysis of Golgi and soluble Golgi fractions identified up-regulated proteins involved in platelet-endothelial adhesion, phospholipase activity, and IFN activity. Thirty four rickettsial proteins were identified with high confidence in the Golgi, plasma membrane, or secreted protein fractions. The host proteins associated with rickettsial infections indicate activation of interferon-STAT signaling pathways; the disruption of cellular adhesion and alteration of antigen presentation pathways in response to rickettsial infections are distinct from those produced by nonspecific LPS stimulation. These patterns of differentially expressed proteins suggest mechanisms of pathogenesis as well as methods for diagnosis and monitoring Rickettsia infections.
© 2016 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures
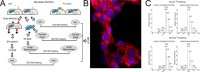
Fig. 1.
Quantitative proteomics study of R. conorii infection in HUVECs. A, experimental strategy. Shown is a schematic diagram of the experimental work flow for the identification of differential protein expression control (LPS-stimulated) or _R. conorii_-infected HUVECs. B, immunofluorescence assay. Immunofluorescence assay for rickettsial antigen (red) and nuclear DNA (blue) in HUVECs smeared on a glass slide to determine rickettsial growth. Original objective magnification was ×40. The primary antibody was a rabbit anti-R. conorii immune serum. The secondary antibody was a donkey anti-rabbit labeled with Alexa 546. C, 18O-labeling efficiency. MS spectra of two 18O-labeled peptides (HSPB1, LATQSNEITIPVTFESR, and ANXA2, GVDEVTIVNILTNR) are shown.
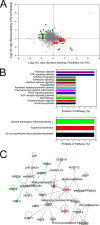
Fig. 2.
Differential protein expression in _R. conorii_-infected HUVEC WCLs. A, quantification of regulation. The plots are log2-transformed forward and reverse heavy/light ratios of individual proteins quantitated in WCLs from each replicate. Up-regulated proteins identified from the _R. conorii-_infected HUVECs are located in the bottom-right quadrant (red circles), and the proteins down-regulated in _Rickettsia_-infected HUVECs are located in the upper-left quadrant (green squares). B, Panther pathway analysis. Top panel, top pathways for proteins enriched in HUVEC WCL identified by the Panther classification system. x axis is the percentage of the pathway represented in the identified proteins. Bottom panel is pathway analysis for proteins depleted in HUVEC WCLs. C, network by IPA. Shown is the top-ranked network of differentially expressed WCL proteins identified in the Ingenuity Knowledge base. For each node, red indicates up-regulation in _Rickettsia-_infected cells; green, down-regulation. For abbreviations, see Table I.
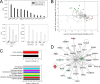
Fig. 3.
Differential protein expression in _R. conorii_-infected HUVEC PM fractions. A, enrichment analysis. Top, spectral count measurements (NSAF) for representative plasma membrane proteins in the PM and WCL fractions. Note the enrichment of plasma membrane in the PM fractions. Bottom left panel, NSAF for cytosolic proteins, which are reduced (depleted) in PM fractions. Bottom right panel, NSAF for representative mitochondrial, endoplasmic reticulum, and nuclear proteins, also depleted in PM fractions. B, quantification of regulation. Shown are up-regulated proteins in PM fractions from _Rickettsia_-infected HUVECs in the bottom-right quadrant (red), and the proteins down-regulated in _Rickettsia_-infected HUVECs are located in the top-left quadrant (green). C, Panther pathway analysis. Top panel, highest ranked pathways identified by the Panther classification system for proteins enriched in HUVEC PMs. Bottom panel, pathways associated with down-regulated PM proteins. D, IPA network. Shown is the top-ranked network (“organismal disease”) of PM proteins in the IPA Knowledge Base. Abbreviations are shown in Table II.
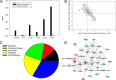
Fig. 4.
Differential protein expression in _R. conorii_-infected HUVEC Golgi fractions. A, enrichment analysis. NSAF for selected Golgi proteins in HUVEC Golgi and WCL fractions. Note the high spectral counts in the Golgi fractions relative to that in WCL fractions. B, quantification of regulation. Golgi proteins up-regulated in _Rickettsia_-stimulated HUVECs are located in the bottom-right quadrant (red), the Golgi proteins down-regulated in Rickettsia stimulated cells are located in the upper left (green). C, Panther pathway analysis. Top panel, top ranked pathways for proteins enriched in HUVEC Golgi fractions. Bottom panel, pathway analysis for proteins depleted in HUVEC Golgi fractions. D, IPA network analysis. Shown is the top-ranked network (“cell-cell signaling and compromise”) of Golgi proteins in IPA. Abbreviations are shown in Table III.
Fig. 5.
Differential expression of secreted proteins in R. conorii infection. A, enrichment analysis. NSAF for selected secreted proteins in soluble Golgi fraction relative to WCL fractions. Note the high spectral counts in the soluble fractions relative to that in WCLs. B, quantification of regulation. Secreted proteins identified in soluble Golgi fraction. Proteins up-regulated in _Rickettsia_-infected HUVECs are located in the bottom-right quadrant (red), and down-regulated proteins are located in the upper left (green). C, Panther protein classification. Shown are the protein classifications for the secreted proteins in the _R. conorii_-infected HUVECs. D, IPA network. Shown is the top-ranked network (“immunological disease”) of soluble Golgi proteins from IPA. Abbreviations are shown in Table IV.
Fig. 6.
Qualification of innate response proteins in _R. conorii_-infected HUVECs. A, qualification of IFN pathway. Shown are SID-selected reaction-monitoring (SRM)-MS measurements of STAT1 and ISG15 (bottom) for HUVEC stimulated with LPS (left) or infected with R. conorii (right). y axis is ratio of protein relative to internal SIS peptide (native/SIS) peptide (native/aqua). B, qualification of rickettsial proteins in Golgi membranes. SID-SRM-MS measurements for UvrABC system protein C are shown; putative ankyrin repeat protein RBE; and chaperone protein HtpG. Compared with LPS-stimulated cells, significant induction of each was observed. C, qualification of rickettsial proteins in soluble Golgi fraction. SID-SRM-MS measurements for human HLA proteins and the rickettsial proteins UVRVC and HPTG in the soluble Golgi fraction.
Similar articles
- Quantitative Proteomics of the Endothelial Secretome Identifies RC0497 as Diagnostic of Acute Rickettsial Spotted Fever Infections.
Zhao Y, Fang R, Zhang J, Zhang Y, Bechelli J, Smalley C, Valbuena G, Walker DH, Oteo JA, Brasier AR. Zhao Y, et al. Am J Pathol. 2020 Feb;190(2):306-322. doi: 10.1016/j.ajpath.2019.10.007. Epub 2020 Jan 16. Am J Pathol. 2020. PMID: 31955791 Free PMC article. - Beta interferon-mediated activation of signal transducer and activator of transcription protein 1 interferes with Rickettsia conorii replication in human endothelial cells.
Colonne PM, Eremeeva ME, Sahni SK. Colonne PM, et al. Infect Immun. 2011 Sep;79(9):3733-43. doi: 10.1128/IAI.05008-11. Epub 2011 Jun 20. Infect Immun. 2011. PMID: 21690236 Free PMC article. - Suppressor of cytokine signalling protein SOCS1 and UBP43 regulate the expression of type I interferon-stimulated genes in human microvascular endothelial cells infected with Rickettsia conorii.
Colonne PM, Sahni A, Sahni SK. Colonne PM, et al. J Med Microbiol. 2013 Jul;62(Pt 7):968-979. doi: 10.1099/jmm.0.054502-0. Epub 2013 Apr 4. J Med Microbiol. 2013. PMID: 23558133 Free PMC article. - Janus Kinase Inhibitors in the Treatment of Vitiligo: A Review.
Qi F, Liu F, Gao L. Qi F, et al. Front Immunol. 2021 Nov 18;12:790125. doi: 10.3389/fimmu.2021.790125. eCollection 2021. Front Immunol. 2021. PMID: 34868078 Free PMC article. Review.
Cited by
- Mediterranean Spotted Fever: Current Knowledge and Recent Advances.
Spernovasilis N, Markaki I, Papadakis M, Mazonakis N, Ierodiakonou D. Spernovasilis N, et al. Trop Med Infect Dis. 2021 Sep 24;6(4):172. doi: 10.3390/tropicalmed6040172. Trop Med Infect Dis. 2021. PMID: 34698275 Free PMC article. Review. - Group B streptococcus exploits vaginal epithelial exfoliation for ascending infection.
Vornhagen J, Armistead B, Santana-Ufret V, Gendrin C, Merillat S, Coleman M, Quach P, Boldenow E, Alishetti V, Leonhard-Melief C, Ngo LY, Whidbey C, Doran KS, Curtis C, Waldorf KMA, Nance E, Rajagopal L. Vornhagen J, et al. J Clin Invest. 2018 May 1;128(5):1985-1999. doi: 10.1172/JCI97043. Epub 2018 Apr 9. J Clin Invest. 2018. PMID: 29629904 Free PMC article. - Multi-omics Analysis Sheds Light on the Evolution and the Intracellular Lifestyle Strategies of Spotted Fever Group Rickettsia spp.
El Karkouri K, Kowalczewska M, Armstrong N, Azza S, Fournier PE, Raoult D. El Karkouri K, et al. Front Microbiol. 2017 Jul 20;8:1363. doi: 10.3389/fmicb.2017.01363. eCollection 2017. Front Microbiol. 2017. PMID: 28775717 Free PMC article. - Revisiting Ehrlichia ruminantium Replication Cycle Using Proteomics: The Host and the Bacterium Perspectives.
Marcelino I, Holzmuller P, Coelho A, Mazzucchelli G, Fernandez B, Vachiéry N. Marcelino I, et al. Microorganisms. 2021 May 26;9(6):1144. doi: 10.3390/microorganisms9061144. Microorganisms. 2021. PMID: 34073568 Free PMC article. - Quantitative Proteomics of the Endothelial Secretome Identifies RC0497 as Diagnostic of Acute Rickettsial Spotted Fever Infections.
Zhao Y, Fang R, Zhang J, Zhang Y, Bechelli J, Smalley C, Valbuena G, Walker DH, Oteo JA, Brasier AR. Zhao Y, et al. Am J Pathol. 2020 Feb;190(2):306-322. doi: 10.1016/j.ajpath.2019.10.007. Epub 2020 Jan 16. Am J Pathol. 2020. PMID: 31955791 Free PMC article.
References
- Walker D. H., Valbuena G. A., and Olano J. P. (2003) Pathogenic mechanisms of diseases caused by Rickettsia. Ann. N.Y. Acad. Sci. 990, 1–11 - PubMed
- Martinez J. J., and Cossart P. (2004) Early signaling events involved in the entry of Rickettsia conorii into mammalian cells. J. Cell Sci. 117, 5097–5106 - PubMed
- Renesto P., Dehoux P., Gouin E., Touqui L., Cossart P., and Raoult D. (2003) Identification and characterization of a phospholipase D-superfamily gene in rickettsiae. J. Infect. Dis. 188, 1276–1283 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- HHSN272200800048C/AI/NIAID NIH HHS/United States
- P30 ES006676/ES/NIEHS NIH HHS/United States
- UL1 TR000071/TR/NCATS NIH HHS/United States
- UL1TR000071/TR/NCATS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous