TGS-TB: Total Genotyping Solution for Mycobacterium tuberculosis Using Short-Read Whole-Genome Sequencing - PubMed (original) (raw)
TGS-TB: Total Genotyping Solution for Mycobacterium tuberculosis Using Short-Read Whole-Genome Sequencing
Tsuyoshi Sekizuka et al. PLoS One. 2015.
Abstract
Whole-genome sequencing (WGS) with next-generation DNA sequencing (NGS) is an increasingly accessible and affordable method for genotyping hundreds of Mycobacterium tuberculosis (Mtb) isolates, leading to more effective epidemiological studies involving single nucleotide variations (SNVs) in core genomic sequences based on molecular evolution. We developed an all-in-one web-based tool for genotyping Mtb, referred to as the Total Genotyping Solution for TB (TGS-TB), to facilitate multiple genotyping platforms using NGS for spoligotyping and the detection of phylogenies with core genomic SNVs, IS6110 insertion sites, and 43 customized loci for variable number tandem repeat (VNTR) through a user-friendly, simple click interface. This methodology is implemented with a KvarQ script to predict MTBC lineages/sublineages and potential antimicrobial resistance. Seven Mtb isolates (JP01 to JP07) in this study showing the same VNTR profile were accurately discriminated through median-joining network analysis using SNVs unique to those isolates. An additional IS6110 insertion was detected in one of those isolates as supportive genetic information in addition to core genomic SNVs. The results of in silico analyses using TGS-TB are consistent with those obtained using conventional molecular genotyping methods, suggesting that NGS short reads could provide multiple genotypes to discriminate multiple strains of Mtb, although longer NGS reads (≥ 300-mer) will be required for full genotyping on the TGS-TB web site. Most available short reads (~100-mer) can be utilized to discriminate the isolates based on the core genome phylogeny. TGS-TB provides a more accurate and discriminative strain typing for clinical and epidemiological investigations; NGS strain typing offers a total genotyping solution for Mtb outbreak and surveillance. TGS-TB web site: https://gph.niid.go.jp/tgs-tb/.
Conflict of interest statement
Competing Interests: The authors have declared that no competing interests exist.
Figures
Fig 1. Schematic representation of the TGS-TB system.
Fig 2. Sample results obtained from TGS-TB.
The NGS reads of seven Mtb isolates were investigated, and the resulting basic information, such as number of trimmed map reads and the coverage region-depth, is shown. In total, 21,805 core-genome SNVs are available in the TGS-TB; 20,928 (95.98%) SNVs are characterized, and 219 additional strain-specific SNVs sites can be implemented in the original dataset. The respective results for lineage, AMR, core genome phylogenetic tree (maximum-likelihood method with x100 bootstrapping), spoligotyping, IS_6110_ insertion and sMIRU-VNTR typing can be viewed in a new window tab and retrieved using the “download all” button. The KvarQ script predicts the lineages/sublineages and AMRs, and the sample queries are assigned as a lineage 2/Beijing sublineage without AMRs (S2 Fig). The AMR target list in the original KvarQ (v2.0) program has been improved with the addition of more reliable genetic alterations for the embA, gyrA, katG, pncA, rpoB, rpsL, rrs and inhA genes (S1 Text).
Fig 3. The core genome phylogeny obtained by the maximum-likelihood method with x100 bootstrapping.
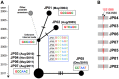
Fig 4. Median-joining network of the seven outbreak isolates based on the detected core genomic variations.
A) The variations are summarized as nexus format files (.nex), and PopART visualizes the epidemiological linkages among the isolates through a user specified network method. The bars on the edge indicate the number of SNVs between the nodes (isolates). B) In addition to three SNV differences between JP05 and the outbreak isolates (JP03, JP04, JP06, and JP07), an additional IS_6110_ insertion was detected at the 1,531,598 nt genome position in JP05, suggesting that JP05 could be unrelated to the outbreak, although the VNTR profile is consistent.
Fig 5. Schematic representation of the IS_6110_ insertion detection strategy.
A) The IS_6110_ sequence (Acc.# X94955 and X94956)-positive short reads are collected (A1-2), rearranged (A3), trimmed (A4), subtracted (A5) and mapped to the Mtb H37Rv chromosome (NC_000962.3) [30] using BWA-SW mapping [29]. B) Typical read mapping profile for the detection of the IS_6110_ insertion site in both directions.
Similar articles
- [Future prospects of molecular epidemiology in tuberculosis].
Matsumoto T, Iwamoto T. Matsumoto T, et al. Kekkaku. 2009 Dec;84(12):783-4. Kekkaku. 2009. PMID: 20077862 Japanese. - The Evolution of Strain Typing in the Mycobacterium tuberculosis Complex.
Merker M, Kohl TA, Niemann S, Supply P. Merker M, et al. Adv Exp Med Biol. 2017;1019:43-78. doi: 10.1007/978-3-319-64371-7_3. Adv Exp Med Biol. 2017. PMID: 29116629 Review. - [New era in molecular epidemiology of tuberculosis in Japan].
Takashima T, Iwamoto T. Takashima T, et al. Kekkaku. 2006 Nov;81(11):693-707. Kekkaku. 2006. PMID: 17154049 Review. Japanese. - DNA fingerprinting of Mycobacterium tuberculosis: from phage typing to whole-genome sequencing.
Schürch AC, van Soolingen D. Schürch AC, et al. Infect Genet Evol. 2012 Jun;12(4):602-9. doi: 10.1016/j.meegid.2011.08.032. Epub 2011 Nov 3. Infect Genet Evol. 2012. PMID: 22067515 - Genotyping of Mycobacterium tuberculosis spreading in Hanoi, Vietnam using conventional and whole genome sequencing methods.
Maeda S, Hijikata M, Hang NTL, Thuong PH, Huan HV, Hoang NP, Hung NV, Cuong VC, Miyabayashi A, Seto S, Keicho N. Maeda S, et al. Infect Genet Evol. 2020 Mar;78:104107. doi: 10.1016/j.meegid.2019.104107. Epub 2019 Nov 6. Infect Genet Evol. 2020. PMID: 31706080
Cited by
- MIRU-profiler: a rapid tool for determination of 24-loci MIRU-VNTR profiles from assembled genomes of Mycobacterium tuberculosis.
Rajwani R, Shehzad S, Siu GKH. Rajwani R, et al. PeerJ. 2018 Jul 11;6:e5090. doi: 10.7717/peerj.5090. eCollection 2018. PeerJ. 2018. PMID: 30018852 Free PMC article. - A step-by-step beginner's protocol for whole genome sequencing of human bacterial pathogens.
Gautam SS, Kc R, Leong KW, Mac Aogáin M, O'Toole RF. Gautam SS, et al. J Biol Methods. 2019 Mar 15;6(1):e110. doi: 10.14440/jbm.2019.276. eCollection 2019. J Biol Methods. 2019. PMID: 31453259 Free PMC article. - Discovery of a unique Mycobacterium tuberculosis protein through proteomic analysis of urine from patients with active tuberculosis.
Pollock N, Dhiman R, Daifalla N, Farhat M, Campos-Neto A. Pollock N, et al. Microbes Infect. 2018 Apr;20(4):228-235. doi: 10.1016/j.micinf.2017.12.011. Epub 2018 Jan 3. Microbes Infect. 2018. PMID: 29306028 Free PMC article. - Overcoming the pitfalls of automatic interpretation of whole genome sequencing data by online tools for the prediction of pyrazinamide resistance in Mycobacterium tuberculosis.
Iwamoto T, Murase Y, Yoshida S, Aono A, Kuroda M, Sekizuka T, Yamashita A, Kato K, Takii T, Arikawa K, Kato S, Mitarai S. Iwamoto T, et al. PLoS One. 2019 Feb 28;14(2):e0212798. doi: 10.1371/journal.pone.0212798. eCollection 2019. PLoS One. 2019. PMID: 30817803 Free PMC article. - Genetic diversity of Mycobacterium tuberculosis isolates from Tochigi prefecture, a local region of Japan.
Mizukoshi F, Miyoshi-Akiyama T, Iwai H, Suzuki T, Kiritani R, Kirikae T, Funatogawa K. Mizukoshi F, et al. BMC Infect Dis. 2017 May 25;17(1):365. doi: 10.1186/s12879-017-2457-y. BMC Infect Dis. 2017. PMID: 28545488 Free PMC article.
References
- Global tuberculosis report. [Internet]. http://www.who.int/tb/publications/global_report/gtbr13_main_text.pdf (accessed Jan 10, 2014). 2013
- Kremer K, van Soolingen D, Frothingham R, Haas WH, Hermans PW, Martin C, et al. Comparison of methods based on different molecular epidemiological markers for typing of Mycobacterium tuberculosis complex strains: interlaboratory study of discriminatory power and reproducibility. Journal of clinical microbiology. 1999;37(8):2607–18. - PMC - PubMed
Publication types
MeSH terms
Grants and funding
This research was funded through a Grant-in-Aid for Research on Emerging and Re-emerging Infectious Diseases (H25-Shinko-Ippan-015) from the Ministry of Health Labour and Welfare Programs of Japan, and also through Research Program on Emerging and Re-emerging Infectious Diseases (15fk0108011h0003 and 15fk0108004h0001) from Japan Agency for Medical Research and development, AMED. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous