The casposon-encoded Cas1 protein from Aciduliprofundum boonei is a DNA integrase that generates target site duplications - PubMed (original) (raw)
. 2015 Dec 15;43(22):10576-87.
doi: 10.1093/nar/gkv1180. Epub 2015 Nov 16.
Affiliations
- PMID: 26573596
- PMCID: PMC4678821
- DOI: 10.1093/nar/gkv1180
The casposon-encoded Cas1 protein from Aciduliprofundum boonei is a DNA integrase that generates target site duplications
Alison B Hickman et al. Nucleic Acids Res. 2015.
Abstract
Many archaea and bacteria have an adaptive immune system known as CRISPR which allows them to recognize and destroy foreign nucleic acid that they have previously encountered. Two CRISPR-associated proteins, Cas1 and Cas2, are required for the acquisition step of adaptation, in which fragments of foreign DNA are incorporated into the host CRISPR locus. Cas1 genes have also been found scattered in several archaeal and bacterial genomes, unassociated with CRISPR loci or other cas proteins. Rather, they are flanked by nearly identical inverted repeats and enclosed within direct repeats, suggesting that these genetic regions might be mobile elements ('casposons'). To investigate this possibility, we have characterized the in vitro activities of the putative Cas1 transposase ('casposase') from Aciduliprofundum boonei. The purified Cas1 casposase can integrate both short oligonucleotides with inverted repeat sequences and a 2.8 kb excised mini-casposon into target DNA. Casposon integration occurs without target specificity and generates 14-15 basepair target site duplications, consistent with those found in casposon host genomes. Thus, Cas1 casposases carry out similar biochemical reactions as the CRISPR Cas1-Cas2 complex but with opposite substrate specificities: casposases integrate specific sequences into random target sites, whereas CRISPR Cas1-Cas2 integrates essentially random sequences into a specific site in the CRISPR locus.
Published by Oxford University Press on behalf of Nucleic Acids Research 2015. This work is written by (a) US Government employee(s) and is in the public domain in the US.
Figures
Figure 1.
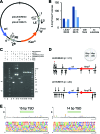
Features of casposon-encoded Cas1 family 2 casposons. (A) The 9075 bp Aciduliprofundum boonei casposon encodes six predicted proteins annotated by NCBI and (18) as: a DNA polymerase type B (red; bp 380 359–383 058); an HNH endonuclease (green; bp 383 051–383 485); a hypothetical protein (beige; bp 383 492–384 064) containing an HTH domain; a transcriptional regulator containing an HTH domain (orange; bp 384 136–385 170); a Cas1 protein (blue; bp 385 171–386 385); and an N6-methyltransferase (yellow; bp 386 408–389 311). The bp numbering is that of the complete genome of Aciduliprofundum boonei T469 (Accession number NC_013926.1). (B) DNA sequence alignment of the TSD-TIR junctions for seven members of casposon-encoded Cas1 family 2. The abbreviations are: MetFor, Methanoregula formicicum SMSP (Accession number NC_019943); MetMaz, Methanosarcina mazei Goe1 (NC_003901); MetMah, Methanohalophilus mahii DSM 5219 (NC_014002); MetTin, Methanolobus tindarius DSM 2278 (NZ_AZAJ01000001); MetLum, Methanomassiliicoccus luminyensis B10 (NZ_CAJE01000015); AciBoo, Aciduliprofundum boonei T469 (NC_013926.1); MetArv, Methanocella arvoryzae MRE50 (NC_009464). Only the top strand sequence is shown. The conserved G/C motif at each end is boxed in grey, the target site duplications (TSDs) are shown in green, and notable sequence patterns are highlighted in red or blue. Bases shown in bold are those that differ between the LE and RE TIRs of a given casposon. The perfect palindrome in the terminal 20 bp of the AciBoo RE is underlined (there is a single bp difference compared to the LE). (C) Amino acid sequence alignment (using T-Coffee) of the Cas1 C-terminal helix-turn-helix (HTH) domains of seven members of the casposon-encoded Cas1 family 2. Only six residues are completely conserved within this alignment (boxed in grey, asterisk below the alignment), yet there are notable regions of identity between subsets of proteins, highlighted in red or blue. The double dots below the alignment represent conservative mutations; single dots are semi-conservative mutations. The α-helices of the HTH domain were predicted using Jpred4 (49). The number at the end of each sequence indicates the number of amino acids in each protein.
Figure 2.
Predicted secondary structure elements of A. boonei Cas1 and alignment with three structurally characterized CRISPR-Cas1 proteins. The β-strands (red arrows) and α-helices (blue cylinders) for Ab Cas1 were predicted using JPred4 (49). The structure-based alignment of Cas1 proteins from E. coli (PDB ID 4P6I), P. aeruginosa (3GOD), and A. fulgidus (4N06) does not differ substantively from those previously reported (9,12,18). The grey secondary structure elements shown below the alignment are those of A. fulgidus. Strictly conserved residues in this alignment are highlighted in grey, and active site residues are boxed. Active site residue numbers are those of Ab Cas1.
Figure 3.
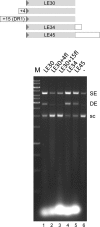
Characterization of the in vitro strand transfer activity of A. boonei Cas1. (A) Strand transfer assay using 30-mer oligonucleotides representing the LE TIR (LE30) or a random sequence (ran30). Insertion of a single-end (SE) results in a relaxed target plasmid, whereas double-end insertion (DE) results in a linearized plasmid. Standard reaction conditions were used with the variations as indicated. In all figures, ‘M’ refers to lanes containing DNA markers, ‘sc’ indicates supercoiled pUC19. (B) Strand transfer assay using a fluorescently-labelled oligonucleotide to confirm integration. Oligonucleotide used: Lane1 = none; Lane2 = ran30. Lane3: 6-FAM-LE26. The reactions were run in duplicate and the samples loaded onto the same 1.5% agarose gel which was cut in half for analysis by ethidium bromide staining (left) or fluorescence (right). (C) Strand transfer assay with modified LE substrates (sequences are listed in Table 1). LE26, LE21, LE15 represent the length of the LE TIR oligonucleotide used, and ran26, ran21, ran15 are length-matched random oligonucleotides. (D) Time course of SE and DE product formation using a LE30 oligonucleotide substrate.
Figure 4.
In vitro integration of mini-casposons by A. boonei Cas1. (A) Schematic representation of plasmids used as templates for the PCR amplification of mini-casposon substrates. The sequences for Primers 1–4 are listed in Table 1. LE: Left End, RE: Right End, DR: Direct repeat. The restriction site for PmeI is indicated. (B) Number of AmpR + KanR colonies recovered when different mini-casposons were used as a substrate for integration into pUC19. For the mini-casposon with flank present, the results for DR2 are shown. Standard assay conditions were used except the reactions were overnight at 37°C. Reactions were performed in triplicate (indicated in different shades of blue), and the number of colonies for ‘with flank’ were 0, 3 and 2; for ‘no substrate’ were 0, 0 and 0. (C) Restriction digest analysis of plasmid products. One representative plasmid is shown for the LE30/RE30 (Lane 5) and LE15/RE15 (Lane 6) reaction. The mini-casposon (Lane 3; LE15/RE15) is cut by PmeI at pHL2777 nt2753 nt (Lane 2; products marked with arrows on left side) but not by XmnI (Lane 1). pUC19 (Lane 4) is cut by XmnI (Lane 7) but not PmeI (Lane 8). The products of the integration reaction (Lanes 5,6) are cut by both enzymes (Lanes 9–12) generating linearized products of ∼5500 kb. (D) Location of integration sites into pUC19. Direction of the arrows indicate the direction of insertion of the terminal C 3′-OH of the LE bottom strand into the plus strand or the minus strand. The arrow marked with the red asterisk indicates the integration event from which the TSD sequence was subsequently used as DR2. (E) Weblogo (50) for the experimentally obtained TSD sequences. Insertions into the minus strand were reverse complemented before being included in the alignment. Data for the LE30/RE30 and LE15/RE15 mini-casposons were combined.
Figure 5.
Strand transfer of TIR oligonucleotides with flanking DNA-TIR junctions. The assay was run under standard conditions but for 40 min. The arrowhead represents the casposon end. Nucleotides in the flanking sequence used correspond to those of DR1 (see Materials and Methods), and the oligonucleotide sequences are listed in Table 1.
Similar articles
- Casposons - silent heroes of the CRISPR-Cas systems evolutionary history.
Smaruj P, Kieliszek M. Smaruj P, et al. EXCLI J. 2023 Jan 5;22:70-83. doi: 10.17179/excli2022-5581. eCollection 2023. EXCLI J. 2023. PMID: 36814855 Free PMC article. Review. - Casposase structure and the mechanistic link between DNA transposition and spacer acquisition by CRISPR-Cas.
Hickman AB, Kailasan S, Genzor P, Haase AD, Dyda F. Hickman AB, et al. Elife. 2020 Jan 8;9:e50004. doi: 10.7554/eLife.50004. Elife. 2020. PMID: 31913120 Free PMC article. - Sequence motifs recognized by the casposon integrase of Aciduliprofundum boonei.
Béguin P, Chekli Y, Sezonov G, Forterre P, Krupovic M. Béguin P, et al. Nucleic Acids Res. 2019 Jul 9;47(12):6386-6395. doi: 10.1093/nar/gkz447. Nucleic Acids Res. 2019. PMID: 31114911 Free PMC article. - Casposon integration shows strong target site preference and recapitulates protospacer integration by CRISPR-Cas systems.
Béguin P, Charpin N, Koonin EV, Forterre P, Krupovic M. Béguin P, et al. Nucleic Acids Res. 2016 Dec 1;44(21):10367-10376. doi: 10.1093/nar/gkw821. Epub 2016 Sep 20. Nucleic Acids Res. 2016. PMID: 27655632 Free PMC article. - Casposons: mobile genetic elements that gave rise to the CRISPR-Cas adaptation machinery.
Krupovic M, Béguin P, Koonin EV. Krupovic M, et al. Curr Opin Microbiol. 2017 Aug;38:36-43. doi: 10.1016/j.mib.2017.04.004. Epub 2017 May 1. Curr Opin Microbiol. 2017. PMID: 28472712 Free PMC article. Review.
Cited by
- History of CRISPR-Cas from Encounter with a Mysterious Repeated Sequence to Genome Editing Technology.
Ishino Y, Krupovic M, Forterre P. Ishino Y, et al. J Bacteriol. 2018 Mar 12;200(7):e00580-17. doi: 10.1128/JB.00580-17. Print 2018 Apr 1. J Bacteriol. 2018. PMID: 29358495 Free PMC article. Review. - Recent Mobility of Casposons, Self-Synthesizing Transposons at the Origin of the CRISPR-Cas Immunity.
Krupovic M, Shmakov S, Makarova KS, Forterre P, Koonin EV. Krupovic M, et al. Genome Biol Evol. 2016 Jan 13;8(2):375-86. doi: 10.1093/gbe/evw006. Genome Biol Evol. 2016. PMID: 26764427 Free PMC article. - Structural basis of seamless excision and specific targeting by piggyBac transposase.
Chen Q, Luo W, Veach RA, Hickman AB, Wilson MH, Dyda F. Chen Q, et al. Nat Commun. 2020 Jul 10;11(1):3446. doi: 10.1038/s41467-020-17128-1. Nat Commun. 2020. PMID: 32651359 Free PMC article. - Casposons - silent heroes of the CRISPR-Cas systems evolutionary history.
Smaruj P, Kieliszek M. Smaruj P, et al. EXCLI J. 2023 Jan 5;22:70-83. doi: 10.17179/excli2022-5581. eCollection 2023. EXCLI J. 2023. PMID: 36814855 Free PMC article. Review. - Using bioinformatic and phylogenetic approaches to classify transposable elements and understand their complex evolutionary histories.
Arkhipova IR. Arkhipova IR. Mob DNA. 2017 Dec 6;8:19. doi: 10.1186/s13100-017-0103-2. eCollection 2017. Mob DNA. 2017. PMID: 29225705 Free PMC article. Review.
References
- Westra E.R., Swarts D.C., Staals R.H.J., Jore M.M., Brouns S.J.J., van der Oost J. The CRISPRs, they are a-changing: how prokaryotes generate adaptive immunity. Annu. Rev. Genet. 2012;46:311–339. - PubMed
- Fineran P.C., Charpentier E. Memory of viral infections by CRISPR-Cas adaptive immune systems: acquisition of new information. Virology. 2012;434:202–209. - PubMed