ff14SB: Improving the Accuracy of Protein Side Chain and Backbone Parameters from ff99SB - PubMed (original) (raw)
ff14SB: Improving the Accuracy of Protein Side Chain and Backbone Parameters from ff99SB
James A Maier et al. J Chem Theory Comput. 2015.
Abstract
Molecular mechanics is powerful for its speed in atomistic simulations, but an accurate force field is required. The Amber ff99SB force field improved protein secondary structure balance and dynamics from earlier force fields like ff99, but weaknesses in side chain rotamer and backbone secondary structure preferences have been identified. Here, we performed a complete refit of all amino acid side chain dihedral parameters, which had been carried over from ff94. The training set of conformations included multidimensional dihedral scans designed to improve transferability of the parameters. Improvement in all amino acids was obtained as compared to ff99SB. Parameters were also generated for alternate protonation states of ionizable side chains. Average errors in relative energies of pairs of conformations were under 1.0 kcal/mol as compared to QM, reduced 35% from ff99SB. We also took the opportunity to make empirical adjustments to the protein backbone dihedral parameters as compared to ff99SB. Multiple small adjustments of φ and ψ parameters were tested against NMR scalar coupling data and secondary structure content for short peptides. The best results were obtained from a physically motivated adjustment to the φ rotational profile that compensates for lack of ff99SB QM training data in the β-ppII transition region. Together, these backbone and side chain modifications (hereafter called ff14SB) not only better reproduced their benchmarks, but also improved secondary structure content in small peptides and reproduction of NMR χ1 scalar coupling measurements for proteins in solution. We also discuss the Amber ff12SB parameter set, a preliminary version of ff14SB that includes most of its improvements.
Figures
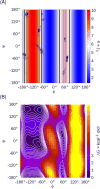
Figure 1
(A) Ramachandran plot with the structures in the ff99SB Ala3 training set shown as circles, with the Hu and Bax H-Hα Karplus curve data shown in the background as a color gradient. Vertical lines indicate the φ values where the Karplus curve matches the scalar coupling value from either NMR (black) or ff99SB simulations (gray). Note that ff99SB training data were limited near the maximum of the Karplus curve (φ=−120°), suggesting that the ff99SB energies may be poorly defined in this region. (B) Free energy surface for alanine dipeptide in ff99SB, showing that the β-ppII transition region near φ,ψ=−120°,160° has significant population despite lack of training data in Figure 1A.
Figure 2
Ramachandran heat maps showing energy differences between ff99SB (lower left, all values 0) and each of the five φ (across) and the four ψ (up) modifications, and all combinations. Note that while these surfaces are graphed with φ and ψ axes, many modifications adjust the φ′ and ψ′ corrections, some with phase shifts, and thus the graphs may not be symmetric about the x and y axes. See main text for definition of “prime” dihedrals.
Figure 3
The AAE of each force field for each amino acid (single letter codes), with data for both α and β backbone conformation. For ionizable residues, the ionic form is indicated by a charge superscript. CC indicates the disulfide bridge. Data are shown for ff99SB, ff99SB-ILDN, and ff99SB with the reparametrized side chain corrections obtained using the procedure described in the text.
Figure 4
RMSD to the NMR structure vs time for the four linear and four native runs of CLN025 with ff14SB and ff99SB, colored by cluster being sampled: black=0, blue=1, green=2, cyan=3, red=4, fuchsia=5, gold=6, and all other clusters light gray.
Figure 5
Average normalized errors (ANE) in side chain scalar couplings for all amino acids in GB3, ubiquitin (Ubq), lysozyme (HEWL), and bovine pancreatic trypsin inhibitor (BPTI), according to ff99SB, ff99SB-ILDN, ff14SBonlysc, and ff14SB. Amino acids are shown with single letter code, with charge state noted for ionizable side chains. Error bars are calculated from four independent simulations.
Figure 6
Order parameters from NMR compared to those back calculated by iRED for ff99SB, ff99SB-ILDN, and ff14SB simulations of GB3, ubiquitin, and lysozyme. Error bars represent the standard deviation of average values from four independent runs. The top panels show differences between simulation and experiment, while the lowest panels show average data for each secondary structure region, following Hornak et al..
Similar articles
- ff19SB: Amino-Acid-Specific Protein Backbone Parameters Trained against Quantum Mechanics Energy Surfaces in Solution.
Tian C, Kasavajhala K, Belfon KAA, Raguette L, Huang H, Migues AN, Bickel J, Wang Y, Pincay J, Wu Q, Simmerling C. Tian C, et al. J Chem Theory Comput. 2020 Jan 14;16(1):528-552. doi: 10.1021/acs.jctc.9b00591. Epub 2019 Dec 3. J Chem Theory Comput. 2020. PMID: 31714766 - Improved side-chain torsion potentials for the Amber ff99SB protein force field.
Lindorff-Larsen K, Piana S, Palmo K, Maragakis P, Klepeis JL, Dror RO, Shaw DE. Lindorff-Larsen K, et al. Proteins. 2010 Jun;78(8):1950-8. doi: 10.1002/prot.22711. Proteins. 2010. PMID: 20408171 Free PMC article. - Comparison of multiple Amber force fields and development of improved protein backbone parameters.
Hornak V, Abel R, Okur A, Strockbine B, Roitberg A, Simmerling C. Hornak V, et al. Proteins. 2006 Nov 15;65(3):712-25. doi: 10.1002/prot.21123. Proteins. 2006. PMID: 16981200 Free PMC article. - The intrinsic conformational features of amino acids from a protein coil library and their applications in force field development.
Jiang F, Han W, Wu YD. Jiang F, et al. Phys Chem Chem Phys. 2013 Mar 14;15(10):3413-28. doi: 10.1039/c2cp43633g. Phys Chem Chem Phys. 2013. PMID: 23385383 - Developments and Applications of Coil-Library-Based Residue-Specific Force Fields for Molecular Dynamics Simulations of Peptides and Proteins.
Jiang F, Wu HN, Kang W, Wu YD. Jiang F, et al. J Chem Theory Comput. 2019 May 14;15(5):2761-2773. doi: 10.1021/acs.jctc.8b00794. Epub 2019 Apr 8. J Chem Theory Comput. 2019. PMID: 30620582 Review.
Cited by
- Cross-Species Susceptibility of Emerging Variants of SARS-CoV-2 Spike.
Li M, Lv F, Li Z, Zhao C, Wang X, Zhu P, Zhou X. Li M, et al. Genes (Basel). 2024 Oct 14;15(10):1321. doi: 10.3390/genes15101321. Genes (Basel). 2024. PMID: 39457447 Free PMC article. - Quantitative Analysis of Protein Unfolded State Energetics: Experimental and Computational Studies Demonstrate That Non-Native Side-Chain Interactions Stabilize Local Native Backbone Structure.
Zou J, Xiao S, Simmerling C, Raleigh DP. Zou J, et al. J Phys Chem B. 2021 Apr 8;125(13):3269-3277. doi: 10.1021/acs.jpcb.0c08922. Epub 2021 Mar 29. J Phys Chem B. 2021. PMID: 33779182 Free PMC article. - Discovery of novel IDO1 inhibitors via structure-based virtual screening and biological assays.
Ge H, Mao L, Zhao J, Wang Y, Shi D, Yang X, Wang X, Liu H, Yao X. Ge H, et al. J Comput Aided Mol Des. 2021 May;35(5):679-694. doi: 10.1007/s10822-021-00386-6. Epub 2021 Apr 27. J Comput Aided Mol Des. 2021. PMID: 33905074 - Histone tails cooperate to control the breathing of genomic nucleosomes.
Huertas J, Schöler HR, Cojocaru V. Huertas J, et al. PLoS Comput Biol. 2021 Jun 3;17(6):e1009013. doi: 10.1371/journal.pcbi.1009013. eCollection 2021 Jun. PLoS Comput Biol. 2021. PMID: 34081696 Free PMC article. - Molecular simulation of SARS-CoV-2 spike protein binding to pangolin ACE2 or human ACE2 natural variants reveals altered susceptibility to infection.
Wang J, Xu X, Zhou X, Chen P, Liang H, Li X, Zhong W, Hao P. Wang J, et al. J Gen Virol. 2020 Sep;101(9):921-924. doi: 10.1099/jgv.0.001452. J Gen Virol. 2020. PMID: 32538738 Free PMC article.
References
- Patel S, Mackerell AD, Brooks CL. CHARMM fluctuating charge force field for proteins: II Protein/solvent properties from molecular dynamics simulations using a nonadditive electrostatic model. J Comp Chem. 2004;25(12):1504–1514. - PubMed
- Patel S, Brooks CL. CHARMM fluctuating charge force field for proteins: I parameterization and application to bulk organic liquid simulations. J Comp Chem. 2004;25(1):1–16. - PubMed
- Anisimov VM, Lamoureux G, Vorobyov IV, Huang N, Roux B, MacKerell AD. Determination of Electrostatic Parameters for a Polarizable Force Field Based on the Classical Drude Oscillator. J Chem Theory Comp. 2005;1(1):153–168. - PubMed
- Lopes PEM, Huang J, Shim J, Luo Y, Li H, Roux B, MacKerell AD. Polarizable Force Field for Peptides and Proteins Based on the Classical Drude Oscillator. J Chem Theory Comp. 2013;9(12):5430–5449. - PMC - PubMed
- Warshel A, Levitt M. Theoretical studies of enzymic reactions: Dielectric, electrostatic and steric stabilization of the carbonium ion in the reaction of lysozyme. J Mol Biol. 1976;103(2):227–249. - PubMed
- Ren P, Ponder JW. Polarizable Atomic Multipole Water Model for Molecular Mechanics Simulation. J Phys Chem B. 2003;107(24):5933–5947.
- Cornell WD, Cieplak P, Bayly CI, Gould IR, Merz KM, Ferguson DM, Spellmeyer DC, Fox T, Caldwell JW, Kollman PA. A Second Generation Force Field For the Simulation of Proteins, Nucleic Acids, and Organic Molecules. J Am Chem Soc. 1995;117(19):5179–5197.
- Wang JM, Cieplak P, Kollman PA. How well does a restrained electrostatic potential (RESP) model perform in calculating conformational energies of organic and biological molecules? J Comp Chem. 2000;21(12):1049–1074.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources