Comparative genomic analyses reveal a vast, novel network of nucleotide-centric systems in biological conflicts, immunity and signaling - PubMed (original) (raw)
. 2015 Dec 15;43(22):10633-54.
doi: 10.1093/nar/gkv1267. Epub 2015 Nov 20.
Affiliations
- PMID: 26590262
- PMCID: PMC4678834
- DOI: 10.1093/nar/gkv1267
Comparative genomic analyses reveal a vast, novel network of nucleotide-centric systems in biological conflicts, immunity and signaling
A Maxwell Burroughs et al. Nucleic Acids Res. 2015.
Abstract
Cyclic di- and linear oligo-nucleotide signals activate defenses against invasive nucleic acids in animal immunity; however, their evolutionary antecedents are poorly understood. Using comparative genomics, sequence and structure analysis, we uncovered a vast network of systems defined by conserved prokaryotic gene-neighborhoods, which encode enzymes generating such nucleotides or alternatively processing them to yield potential signaling molecules. The nucleotide-generating enzymes include several clades of the DNA-polymerase β-like superfamily (including Vibrio cholerae DncV), a minimal version of the CRISPR polymerase and DisA-like cyclic-di-AMP synthetases. Nucleotide-binding/processing domains include TIR domains and members of a superfamily prototyped by Smf/DprA proteins and base (cytokinin)-releasing LOG enzymes. They are combined in conserved gene-neighborhoods with genes for a plethora of protein superfamilies, which we predict to function as nucleotide-sensors and effectors targeting nucleic acids, proteins or membranes (pore-forming agents). These systems are sometimes combined with other biological conflict-systems such as restriction-modification and CRISPR/Cas. Interestingly, several are coupled in mutually exclusive neighborhoods with either a prokaryotic ubiquitin-system or a HORMA domain-PCH2-like AAA+ ATPase dyad. The latter are potential precursors of equivalent proteins in eukaryotic chromosome dynamics. Further, components from these nucleotide-centric systems have been utilized in several other systems including a novel diversity-generating system with a reverse transcriptase. We also found the Smf/DprA/LOG domain from these systems to be recruited as a predicted nucleotide-binding domain in eukaryotic TRPM channels. These findings point to evolutionary and mechanistic links, which bring together CRISPR/Cas, animal interferon-induced immunity, and several other systems that combine nucleic-acid-sensing and nucleotide-dependent signaling.
Published by Oxford University Press on behalf of Nucleic Acids Research 2015. This work is written by (a) US Government employee(s) and is in the public domain in the US.
Figures
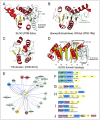
Figure 1.
(A) Genome context network overview of identified systems. Networks constructed as described in Materials and Methods, with blue edges representing domains directly fused in the same polypeptide and gray edges representing links in conserved gene neighborhoods. (B) Network depicting the relationship between nucleotide synthesizing and recognition/processing enzymes (left) with sensor/regulatory domains (middle left), TM-containing domains (middle right) and potential secondary effectors (right). (C–J) Representative depictions of conserved domain architectures and gene neighborhoods containing predicted TM pore-forming effectors (for comprehensive list of architectures and neighborhoods, see Supplementary Material). Domain architectures are depicted as adjacent shapes and are not drawn to scale. Gene neighborhoods are depicted as box arrows. Architectures and neighborhoods are labeled with NCBI gene identifier (gi) number and organism name, separated by underscore. Abbreviation of organism lineage is provided to the right of the label in parenthesis. Abbreviations: prot, proteobacteria; bacter, bacteroidetes; actino, actinobacteria; spiro, spirochaetes; firm, firmicutes; nitro, nitrospirae; cyano, cyanobacteria; plancto, planctomycetes; thermo, thermotogae; aquif, aquifex; tener, tenericutes; fuso, fusobacteria; euryarch, euryarchaea; chlorof, chloroflexi; deino, deinococci; euk, eukaryotes. (K) Network centered on the Patatin hub. (L and M) Network and gene neighborhood representation of newly-identified, preQ0-based R-M system.
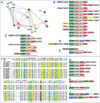
Figure 2.
(A) Cartoon (left) and surface (right) structure rendering of DncV representative of the SMODS family. Active site residues, predicted oligonucleotide recognition residues, and ligand are rendered as ball-and-stick. Predicted recognition residues (left) and surface patch corresponding to R281 side chain (right) are colored in pink. (B–D) Network and representative domain architectures and gene neighborhoods depicting relationships between SAVED and SMODS domains. See Figure 1 legend for abbreviations and further explanation. (E and F) Representative gene neighborhoods depicting nestling of two-gene systems within classical CRISPR/Cas and R-M systems.
Figure 3.
(A–C) Cartoon renderings of SLOG (A), DRHyd (B) and TIR (C) domains. Secondary structure elements specific to SLOG (A) are colored in light blue and rendered as transparent. Strands in the SLOG domain, N- and C-termini, and conserved residues are labeled. (D) Topology diagram of idealized SLOG domain, with strands and helices depicted as boxed arrows and coils, respectively. (E–J) Network and representative gene neighborhoods of systems containing the TIR domain. See Figure 1 for abbreviations and further explanation.
Figure 4.
(A–E) Network and conserved gene neighborhoods representing systems containing the Ubl conjugation systems combined with two-gene systems. See Figure 1 for abbreviations and further explanation. (F) Multiple sequence alignment of bacterial HORMA domains. Sequences in alignment are labeled to the left by organism abbreviation and gi number, separated by underscore. The first three sequences are known eukaryotic HORMA domains with experimentally determined structures labeled by Protein Databank (pdb) id and chain letter, separated by underscore. Bottom line provides amino acid residue consensus conservation for individual columns in the alignment, color-coordinated as follows: o, hydroxyl-bearing colored in orange; s, small colored in green; p, polar colored in blue; h, hydrophobic colored in yellow; b, big colored in gray; u, tiny colored in green. Conserved tryptophan crucial to the conserved HORMA binding pocket is marked with ‘*’ and colored in red. (G–J) Network and representative gene neighborhoods for systems containing the HORMA-TRIP13/Pch2 dyad combined with two-gene systems.
Figure 5.
(A–F) Network and conserved gene neighborhoods representing systems centered on the SLOG domain. See Figure 1 for abbreviations and further explanation. (G) Multiple sequence alignment of SLOG domain family found in TRPM ion and predicted prokaryotic membrane channels (LSDAT family in Table 1). See Figure 4 for explanation of multiple sequence alignment and abbreviations. Conserved residues are colored in red with letters shaded in white. Additional abbreviations: -, negatively-charged residue; l, aliphatic residue. (H) Representative domain architectures of the SLOG domain in TRPM ion channels and other eukaryote contexts. (I) Representative domain architectures of SLOG domains predicted to be part of ‘conventional’ second messenger signaling systems. (J) Representative gene neighborhoods of pJV1-spdB3-based signaling systems. (K) Gene neighborhoods representing predicted signaling switch mediated by SLOG and PRTase domains.
Similar articles
- Identification of Uncharacterized Components of Prokaryotic Immune Systems and Their Diverse Eukaryotic Reformulations.
Burroughs AM, Aravind L. Burroughs AM, et al. J Bacteriol. 2020 Nov 19;202(24):e00365-20. doi: 10.1128/JB.00365-20. Print 2020 Nov 19. J Bacteriol. 2020. PMID: 32868406 Free PMC article. - Comparative genomics of the FtsK-HerA superfamily of pumping ATPases: implications for the origins of chromosome segregation, cell division and viral capsid packaging.
Iyer LM, Makarova KS, Koonin EV, Aravind L. Iyer LM, et al. Nucleic Acids Res. 2004 Oct 5;32(17):5260-79. doi: 10.1093/nar/gkh828. Print 2004. Nucleic Acids Res. 2004. PMID: 15466593 Free PMC article. - Polymorphic toxin systems: Comprehensive characterization of trafficking modes, processing, mechanisms of action, immunity and ecology using comparative genomics.
Zhang D, de Souza RF, Anantharaman V, Iyer LM, Aravind L. Zhang D, et al. Biol Direct. 2012 Jun 25;7:18. doi: 10.1186/1745-6150-7-18. Biol Direct. 2012. PMID: 22731697 Free PMC article. - Regulation of CRISPR-Cas adaptive immune systems.
Patterson AG, Yevstigneyeva MS, Fineran PC. Patterson AG, et al. Curr Opin Microbiol. 2017 Jun;37:1-7. doi: 10.1016/j.mib.2017.02.004. Epub 2017 Mar 27. Curr Opin Microbiol. 2017. PMID: 28359988 Review. - The many faces of the helix-turn-helix domain: transcription regulation and beyond.
Aravind L, Anantharaman V, Balaji S, Babu MM, Iyer LM. Aravind L, et al. FEMS Microbiol Rev. 2005 Apr;29(2):231-62. doi: 10.1016/j.femsre.2004.12.008. FEMS Microbiol Rev. 2005. PMID: 15808743 Review.
Cited by
- SEN1990 is a predicted winged helix-turn-helix protein involved in the pathogenicity of Salmonella enterica serovar Enteritidis and the expression of the gene oafB in the SPI-17.
Hoppe-Elsholz G, Piña-Iturbe A, Vallejos OP, Suazo ID, Sepúlveda-Alfaro J, Pereira-Sánchez P, Martínez-Balboa Y, Catalán EA, Reyes P, Scaff V, Bassi F, Campos-Gajardo S, Avilés A, Santiviago CA, Kalergis AM, Bueno SM. Hoppe-Elsholz G, et al. Front Microbiol. 2023 Nov 3;14:1236458. doi: 10.3389/fmicb.2023.1236458. eCollection 2023. Front Microbiol. 2023. PMID: 38029095 Free PMC article. - Conservation and similarity of bacterial and eukaryotic innate immunity.
Ledvina HE, Whiteley AT. Ledvina HE, et al. Nat Rev Microbiol. 2024 Jul;22(7):420-434. doi: 10.1038/s41579-024-01017-1. Epub 2024 Feb 28. Nat Rev Microbiol. 2024. PMID: 38418927 Free PMC article. Review. - Ubiquitin-like conjugation by bacterial cGAS enhances anti-phage defence.
Jenson JM, Li T, Du F, Ea CK, Chen ZJ. Jenson JM, et al. Nature. 2023 Apr;616(7956):326-331. doi: 10.1038/s41586-023-05862-7. Epub 2023 Feb 27. Nature. 2023. PMID: 36848932 Free PMC article. - An E1-E2 fusion protein primes antiviral immune signalling in bacteria.
Ledvina HE, Ye Q, Gu Y, Sullivan AE, Quan Y, Lau RK, Zhou H, Corbett KD, Whiteley AT. Ledvina HE, et al. Nature. 2023 Apr;616(7956):319-325. doi: 10.1038/s41586-022-05647-4. Epub 2023 Feb 8. Nature. 2023. PMID: 36755092 Free PMC article. - A structural overview of the ion channels of the TRPM family.
Huang Y, Fliegert R, Guse AH, Lü W, Du J. Huang Y, et al. Cell Calcium. 2020 Jan;85:102111. doi: 10.1016/j.ceca.2019.102111. Epub 2019 Nov 24. Cell Calcium. 2020. PMID: 31812825 Free PMC article. Review.
References
- Sutherland E.W., Rall T.W. Fractionation and characterization of a cyclic adenine ribonucleotide formed by tissue particles. J. Biol. Chem. 1958;232:1077–1091. - PubMed
- Rall T.W., Sutherland E.W. Formation of a cyclic adenine ribonucleotide by tissue particles. J. Biol. Chem. 1958;232:1065–1076. - PubMed
- Gancedo J.M. Biological roles of cAMP: variations on a theme in the different kingdoms of life. Biol. Rev. Camb. Philos. Soc. 2013;88:645–668. - PubMed
- Gorke B., Stulke J. Carbon catabolite repression in bacteria: many ways to make the most out of nutrients. Nat. Rev. Microbiol. 2008;6:613–624. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources