Chromosomal Loop Domains Direct the Recombination of Antigen Receptor Genes - PubMed (original) (raw)
Chromosomal Loop Domains Direct the Recombination of Antigen Receptor Genes
Jiazhi Hu et al. Cell. 2015.
Abstract
RAG initiates antibody V(D)J recombination in developing lymphocytes by generating "on-target" DNA breaks at matched pairs of bona fide recombination signal sequences (RSSs). We employ bait RAG-generated breaks in endogenous or ectopically inserted RSS pairs to identify huge numbers of RAG "off-target" breaks. Such breaks occur at the simple CAC motif that defines the RSS cleavage site and are largely confined within convergent CTCF-binding element (CBE)-flanked loop domains containing bait RSS pairs. Marked orientation dependence of RAG off-target activity within loops spanning up to 2 megabases implies involvement of linear tracking. In this regard, major RAG off-targets in chromosomal translocations occur as convergent RSS pairs at enhancers within a loop. Finally, deletion of a CBE-based IgH locus element disrupts V(D)J recombination domains and, correspondingly, alters RAG on- and off-target distributions within IgH. Our findings reveal how RAG activity is developmentally focused and implicate mechanisms by which chromatin domains harness biological processes within them.
Copyright © 2015 Elsevier Inc. All rights reserved.
Figures
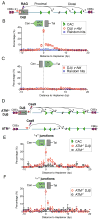
Figure 1. Abundant DSBs in the 1.8-Mb _c-Myc_-DJβ Loop Domain in v-Abl Pro-B Cells
(A) Diagram of _c-My_c-DJβ and sequences of Dβ1 23RSS (blue) and Jβ1-1 12RSS (orange). Red triangles indicate RAG cleavage-sites. (B) RAG-initiated DSBs in _c-Myc_-DJβ cassette participate in cassette DJβ rearrangements but rarely to translocations involving DSBs on other chromosomes. Red arrows indicate HTGTS primer positions. (C) Linear plot with a broken y axis showing HTGTS junction profiles in indicated 20-Mb region containing _c-Myc_-DJβ. (D) Potential junctional outcomes between bait Dβ1 23RSS coding ends and other DSBs in cis include deletions, excision circles, and inversions. (E) HTGTS junction profiles in v-Abl cells within indicated 2-Mb region containing _c-Myc_-DJβ. For all panels, unmarked ticks represent 0. Black lines in the middle show hotspot (HS) positions (listed in Table S1). Junction numbers and percentages in + or − orientation downstream of _c-Myc_-DJβ are shown. Cassette location is shadowed in gray. Star indicates a good cryptic RSS. (F) ChIP-seq profiles of CTCF and Rad21 in the 2-Mb region defined in (E). CBE orientation is indicated by purple triangles. (G) Heat map showing the 1.8-Mb c-Myc loop domain defined by in situ Hi-C data in CH12-LX cell line. See also Figure S1 and Table S1.
Figure 2. RAG Generates DSBs Across the 1.8-Mb _c-Myc_-DJβ Loop Domain
(A) Schematic of translocations between Dβ1 downstream coding ends and cryptic RSSs mostly represented by CAC motifs in the 1.8-Mb c-Myc domain in _c-Myc_-DJβ vAbl pro-B cells. (B, C) Distance of Dβ1 downstream coding end junctions to reverse CACs (B) or forward CACs (C) within the1.8-Mb domain in _c-Myc_-DJβ vAbl pro-B cells. Direct joining to the nucleotide immediately adjacent to CAC is defined as 0 in this and following panels. Mean ± SD, n=3. (D) Schematic of translocations between Cas9/gRNA-initiated bait DSBs and DSBs in the c-Myc domain in ATM-deficient pro-B cells with (upper) or without (lower) the _c-Myc_-DJβ cassette. (E, F) Distance of 5′Cas9 junctions in + (E) or − (F) orientation to reverse CACs in the c-Myc domain in cells defined in (D). Means ± SD, n=3. See also Figure S2.
Figure 3. DSBs Restricted in Genome-wide DEL-SJ-containing Loop Domains
(A) Diagram showing major RAG-initiated joins in DEL-SJ-containing v-Abl pro-B cells. (B) Potential junctional outcomes between bait 12RSS and DSBs in cis include deletions, excision circles, and inversions. (C) Profiles of 12RSS junctions within chromosome X in ATM-deficient vAbl pro-B cells. Black triangles indicate insertion site of DEL-SJ. Junction numbers and percentages in + or − orientation upstream or downstream of bait 12RSS are shown separately. (D, E) Profiles of 23RSS junctions in the indicated 3.5-Mb region containing DEL-SJ on chromosome X. (F) ChIP-seq profiles of CTCF/Rad21 (upper panel) and heat map of in situ Hi-C (lower panel) in this 3.5-Mb region. (G–K) 12RSS and 23RSS junctions across the 1-Mb DEL-SJ-containing loop domain on chromosome 4. Stars indicate stronger cryptic RSSs. See also Figure S3 and Table S2.
Figure 4. Orientation-biased Joining of RAG-initiated DSBs in Loop Domains
(A) Schematic of translocation between bait 12RSS and CACs within the DEL-SJ-containing loop domain on chromosome X in ATM-deficient v-Abl pro-B cells. (B) Distance of 12RSS junctions upstream of bait 12RSS to forward CACs; no correlation was found with reverse CACs. (C) Distance of 12RSS junctions downstream of bait 12RSS to reverse CACs; no correlation was found with forward CACs. (D) Diagram of recombination output generated by RAG re-cleavage at perfect 12–23RSS joins. (E) Profiles of 12RSS junctions of 12–23RSS join within chromosome X in ATM-deficient vAbl pro-B cells. Star indicates a relatively stronger cryptic RSS and loop domain is shadowed in gray, also in (G). (F) Diagram of bait surrogate coding ends associated with DEL-SJ 12RSS and the potential outcomes. (G) Profiles of GFP primer junctions within chromosome X in ATM-deficient vAbl pro-B cells. (H) Distance of GFP primer junctions downstream of bait surrogate coding ends to forward CACs. See also Figure S4.
Figure 5. Genome-wide RAG Off-targets
(A) Circos plot on a custom log scale showing genome-wide translocation profile of GFP primer junctions in DEL-SJ-integrated ATM-deficient v-Abl pro-B cells. Bin size is 2 Mb and 50,000 HTGTS junctions are shown. Colored lines link break-site to identified cryptic RSSs. (B) Consensus sequence of cryptic RSS heptamer extrapolated from the 107 identified cryptic RSSs. (C) Paired cryptic RSSs on chromosome 1 (left panel) and 15 (right panel). Green arrows indicate the position and orientation of cryptic RSSs in these four translocation hotspots. (D) Venn diagram showing number of identified cryptic RSSs that overlap with H3K27Ac and H3K4me3. (E) Pie chart showing number of identified cryptic RSSs that overlap with typical enhancers and super-enhancers. See also Figure S5 and Table S3.
Figure 6. Effect of IGCR1 Deletion on RAG Targeting in the IgH Locus
(A) Schematic of murine IgH locus. Purple arrowheads indicate position and orientation of CBEs. Red arches show proposed looping between convergent IgH CBEs (Lin et al., 2015). (B) Illustration of possible joining outcomes between bona fide RSSs from DFL16.1 5′ RSS bait end to upstream VH and downstream JH broken ends. Red arrows in all panels indicate the position and orientation of HTGTS primer. (C,D) Upper panels are IgH annotation track. Middle panels show CTCF ChIP-seq profiles. Lower panels are pooled HTGTS junction profiles for IgH bona fide RSSs (n=3). Junctions are displayed as stacked tracks (log scale between tracks, linear scale within each track). Junction numbers and percentages as of total IgH on-targets in indicated regions are shown. We note that beyond junctions described in the text we also detected junctions between DFL16.1 5′RSS and pseudo DH RSSs in the VH to DH intervening region in ATM−/−-IGCR1Δ cells (Green star; see also Figure S6E). We also found a very small number of hybrid joins to VH coding ends (+ joins) or JH4 coding end (− joins). (E) Pooled RAG off-target junction profile of a 24-kb region including the recombination domain for ATM-deficient cells (n=3). (F, G) show pooled RAG off-target junction profiles of a 240-kb region including the recombination domain for ATM-deficient cells with (F) or without (G) IGCR1, respectively (n=3). Brown shadowed region marks the location of IGCR1. Junction numbers and percentages as of total IgH off-targets in indicated regions are shown. See also Figure S6 and Table S4.
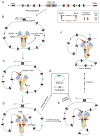
Figure 7. Linear Tracking Model for Orientation-biased Usage of CACs in the Paired bona fide RSSs-containing Loop Domains
(A) Linear map of a DEL-SJ-containing loop domain. (B) Activation of RAG by paired bona fide RSSs in the CTCF/cohesin-anchored loop domain. (C) One bona fide RSS may escape at a very low frequency leaving the activated complex. (D) A CAC in convergent orientation of the remaining bona fide RSS binds to RAG to facilitate cleavage or initiate unidirectional tracking. (E) Further tracking resulting in usage of another convergent CAC. (F) RAG tracking in the loop in either direction is stopped by the boundary formed by the CTCF/cohesin complex. (G) Joining products from pairing and cleavage between remaining bona fide RSS and convergent CACs. More details are in the text. Various modifications of this basic tracking model can be conceived to explain joining patterns of proximal V, D, and J segments in endogenous antigen receptor loci (not shown). See also Figure S7.
Similar articles
- Loop extrusion mediates physiological Igh locus contraction for RAG scanning.
Dai HQ, Hu H, Lou J, Ye AY, Ba Z, Zhang X, Zhang Y, Zhao L, Yoon HS, Chapdelaine-Williams AM, Kyritsis N, Chen H, Johnson K, Lin S, Conte A, Casellas R, Lee CS, Alt FW. Dai HQ, et al. Nature. 2021 Feb;590(7845):338-343. doi: 10.1038/s41586-020-03121-7. Epub 2021 Jan 13. Nature. 2021. PMID: 33442057 Free PMC article. - An Ectopic CTCF Binding Element Inhibits Tcrd Rearrangement by Limiting Contact between Vδ and Dδ Gene Segments.
Chen L, Zhao L, Alt FW, Krangel MS. Chen L, et al. J Immunol. 2016 Oct 15;197(8):3188-3197. doi: 10.4049/jimmunol.1601124. Epub 2016 Sep 9. J Immunol. 2016. PMID: 27613698 Free PMC article. - CTCF-Binding Elements Mediate Accessibility of RAG Substrates During Chromatin Scanning.
Jain S, Ba Z, Zhang Y, Dai HQ, Alt FW. Jain S, et al. Cell. 2018 Jun 28;174(1):102-116.e14. doi: 10.1016/j.cell.2018.04.035. Epub 2018 May 24. Cell. 2018. PMID: 29804837 Free PMC article. - RAG Chromatin Scanning During V(D)J Recombination and Chromatin Loop Extrusion are Related Processes.
Lin SG, Ba Z, Alt FW, Zhang Y. Lin SG, et al. Adv Immunol. 2018;139:93-135. doi: 10.1016/bs.ai.2018.07.001. Epub 2018 Aug 27. Adv Immunol. 2018. PMID: 30249335 Review. - Structural gymnastics of RAG-mediated DNA cleavage in V(D)J recombination.
Ru H, Zhang P, Wu H. Ru H, et al. Curr Opin Struct Biol. 2018 Dec;53:178-186. doi: 10.1016/j.sbi.2018.11.001. Epub 2018 Nov 23. Curr Opin Struct Biol. 2018. PMID: 30476719 Free PMC article. Review.
Cited by
- DNA Damage Response and Repair in Adaptive Immunity.
Luo S, Qiao R, Zhang X. Luo S, et al. Front Cell Dev Biol. 2022 May 17;10:884873. doi: 10.3389/fcell.2022.884873. eCollection 2022. Front Cell Dev Biol. 2022. PMID: 35663402 Free PMC article. Review. - Detecting DNA double-stranded breaks in mammalian genomes by linear amplification-mediated high-throughput genome-wide translocation sequencing.
Hu J, Meyers RM, Dong J, Panchakshari RA, Alt FW, Frock RL. Hu J, et al. Nat Protoc. 2016 May;11(5):853-71. doi: 10.1038/nprot.2016.043. Epub 2016 Mar 31. Nat Protoc. 2016. PMID: 27031497 Free PMC article. - The Chromatin Organization Close to SNP rs12913832, Involved in Eye Color Variation, Is Evolutionary Conserved in Vertebrates.
Brancato D, Bruno F, Coniglio E, Sturiale V, Saccone S, Federico C. Brancato D, et al. Int J Mol Sci. 2024 Jun 15;25(12):6602. doi: 10.3390/ijms25126602. Int J Mol Sci. 2024. PMID: 38928306 Free PMC article. - Highly sensitive and unbiased approach for elucidating antibody repertoires.
Lin SG, Ba Z, Du Z, Zhang Y, Hu J, Alt FW. Lin SG, et al. Proc Natl Acad Sci U S A. 2016 Jul 12;113(28):7846-51. doi: 10.1073/pnas.1608649113. Epub 2016 Jun 27. Proc Natl Acad Sci U S A. 2016. PMID: 27354528 Free PMC article. - The molecular basis and disease relevance of non-homologous DNA end joining.
Zhao B, Rothenberg E, Ramsden DA, Lieber MR. Zhao B, et al. Nat Rev Mol Cell Biol. 2020 Dec;21(12):765-781. doi: 10.1038/s41580-020-00297-8. Epub 2020 Oct 19. Nat Rev Mol Cell Biol. 2020. PMID: 33077885 Free PMC article. Review.
References
- Bossen C, Mansson R, Murre C. Chromatin topology and the regulation of antigen receptor assembly. Annu Rev Immunol. 2012;30:337–356. - PubMed
- Bredemeyer AL, Sharma GG, Huang CY, Helmink BA, Walker LM, Khor KC, Nuskey B, Sullivan KE, Pandita TK, Bassing CH, et al. ATM stabilizes DNA double-strand-break complexes during V(D)J recombination. Nature. 2006;442:466–470. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 AI020047/AI/NIAID NIH HHS/United States
- AI007512/AI/NIAID NIH HHS/United States
- R01 AI032524/AI/NIAID NIH HHS/United States
- AI020047/AI/NIAID NIH HHS/United States
- AI032524/AI/NIAID NIH HHS/United States
- R37 AI020047/AI/NIAID NIH HHS/United States
- T32 AI007512/AI/NIAID NIH HHS/United States
- R37 AI032524/AI/NIAID NIH HHS/United States
- HHMI/Howard Hughes Medical Institute/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases