DNA methylation age is associated with mortality in a longitudinal Danish twin study - PubMed (original) (raw)
DNA methylation age is associated with mortality in a longitudinal Danish twin study
Lene Christiansen et al. Aging Cell. 2016 Feb.
Abstract
An epigenetic profile defining the DNA methylation age (DNAm age) of an individual has been suggested to be a biomarker of aging, and thus possibly providing a tool for assessment of health and mortality. In this study, we estimated the DNAm age of 378 Danish twins, age 30-82 years, and furthermore included a 10-year longitudinal study of the 86 oldest-old twins (mean age of 86.1 at follow-up), which subsequently were followed for mortality for 8 years. We found that the DNAm age is highly correlated with chronological age across all age groups (r = 0.97), but that the rate of change of DNAm age decreases with age. The results may in part be explained by selective mortality of those with a high DNAm age. This hypothesis was supported by a classical survival analysis showing a 35% (4-77%) increased mortality risk for each 5-year increase in the DNAm age vs. chronological age. Furthermore, the intrapair twin analysis revealed a more-than-double mortality risk for the DNAm oldest twin compared to the co-twin and a 'dose-response pattern' with the odds of dying first increasing 3.2 (1.05-10.1) times per 5-year DNAm age difference within twin pairs, thus showing a stronger association of DNAm age with mortality in the oldest-old when controlling for familial factors. In conclusion, our results support that DNAm age qualifies as a biomarker of aging.
Keywords: DNA methylation; biological age; biomarker; epigenetic clock; mortality; twins.
© 2015 The Authors. Aging Cell published by the Anatomical Society and John Wiley & Sons Ltd.
Figures
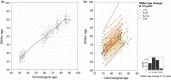
Figure 1
Horvath
DNA
m age against chronological age. (a) Correlation between the methylation age using the Horvath model. The relation is visualized by a smoothing spline. (b)
DNA
m age trajectories of each individual in the longitudinal study of oldest‐olds. The lines are colored by intervals of slow to fast agers, and the histogram indicates the distribution within the intervals.
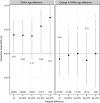
Figure 2
Mortality of the oldest twins according to intrapair
DNA
m age difference at follow‐up and difference in change over 10 years. The plots display the proportion of twins where the
DNA
m oldest twins died first (Proportion dead), for all twins and divided into groups with increasing intrapair difference. The numbers of twin pairs in each group are given in brackets, and the _P_‐values are shown below the lines on the left panel and above the lines on the right.
Similar articles
- Does the epigenetic clock GrimAge predict mortality independent of genetic influences: an 18 year follow-up study in older female twin pairs.
Föhr T, Waller K, Viljanen A, Sanchez R, Ollikainen M, Rantanen T, Kaprio J, Sillanpää E. Föhr T, et al. Clin Epigenetics. 2021 Jun 13;13(1):128. doi: 10.1186/s13148-021-01112-7. Clin Epigenetics. 2021. PMID: 34120642 Free PMC article. - Leisure-time physical activity and DNA methylation age-a twin study.
Sillanpää E, Ollikainen M, Kaprio J, Wang X, Leskinen T, Kujala UM, Törmäkangas T. Sillanpää E, et al. Clin Epigenetics. 2019 Jan 19;11(1):12. doi: 10.1186/s13148-019-0613-5. Clin Epigenetics. 2019. PMID: 30660189 Free PMC article. - Genetic and environmental causes of variation in epigenetic aging across the lifespan.
Li S, Nguyen TL, Wong EM, Dugué PA, Dite GS, Armstrong NJ, Craig JM, Mather KA, Sachdev PS, Saffery R, Sung J, Tan Q, Thalamuthu A, Milne RL, Giles GG, Southey MC, Hopper JL. Li S, et al. Clin Epigenetics. 2020 Oct 22;12(1):158. doi: 10.1186/s13148-020-00950-1. Clin Epigenetics. 2020. PMID: 33092643 Free PMC article. - Do age-associated DNA methylation changes increase the risk of malignant transformation?
Wagner W, Weidner CI, Lin Q. Wagner W, et al. Bioessays. 2015 Jan;37(1):20-4. doi: 10.1002/bies.201400063. Epub 2014 Oct 9. Bioessays. 2015. PMID: 25303747 Review. - Analysis of epigenetic aging in vivo and in vitro: Factors controlling the speed and direction.
Matsuyama M, Søraas A, Yu S, Kim K, Stavrou EX, Caimi PF, Wald D, deLima M, Dahl JA, Horvath S, Matsuyama S. Matsuyama M, et al. Exp Biol Med (Maywood). 2020 Nov;245(17):1543-1551. doi: 10.1177/1535370220947015. Epub 2020 Aug 6. Exp Biol Med (Maywood). 2020. PMID: 32762265 Free PMC article. Review.
Cited by
- Epigenetic age and pregnancy outcomes: GrimAge acceleration is associated with shorter gestational length and lower birthweight.
Ross KM, Carroll JE, Horvath S, Hobel CJ, Coussons-Read ME, Dunkel Schetter C. Ross KM, et al. Clin Epigenetics. 2020 Aug 6;12(1):120. doi: 10.1186/s13148-020-00909-2. Clin Epigenetics. 2020. PMID: 32762768 Free PMC article. - Longitudinal changes and variation in human DNA methylation analysed with the Illumina MethylationEPIC BeadChip assay and their implications on forensic age prediction.
Refn MR, Andersen MM, Kampmann ML, Tfelt-Hansen J, Sørensen E, Larsen MH, Morling N, Børsting C, Pereira V. Refn MR, et al. Sci Rep. 2023 Dec 8;13(1):21658. doi: 10.1038/s41598-023-49064-7. Sci Rep. 2023. PMID: 38066081 Free PMC article. - The "cognitive clock": A novel indicator of brain health.
Boyle PA, Wang T, Yu L, Wilson RS, Dawe R, Arfanakis K, Schneider JA, Beck T, Rajan KB, Evans D, Bennett DA. Boyle PA, et al. Alzheimers Dement. 2021 Dec;17(12):1923-1937. doi: 10.1002/alz.12351. Epub 2021 Jun 1. Alzheimers Dement. 2021. PMID: 34060702 Free PMC article. - Longitudinal Baboon (Papio anubis) Neutrophil to Lymphocyte Ratio (NLR), and Correlations with Monthly Sedation Rate and Within-Group Sedation Order.
Neal SJ, Schapiro SJ, Magden ER. Neal SJ, et al. Vet Sci. 2024 Sep 11;11(9):423. doi: 10.3390/vetsci11090423. Vet Sci. 2024. PMID: 39330802 Free PMC article. - DNA methylation-based variation between human populations.
Kader F, Ghai M. Kader F, et al. Mol Genet Genomics. 2017 Feb;292(1):5-35. doi: 10.1007/s00438-016-1264-2. Epub 2016 Nov 4. Mol Genet Genomics. 2017. PMID: 27815639 Review.
References
- Bell JT, Tsai PC, Yang TP, Pidsley R, Nisbet J, Glass D, Mangino M, Zhai G, Zhang F, Valdes A, Shin SY, Dempster EL, Murray RM, Grundberg E, Hedman AK, Nica A, Small KS, Mu TC, Dermitzakis ET, McCarthy MI, Mill J, Spector TD, Deloukas P (2012) Epigenome‐wide scans identify differentially methylated regions for age and age‐related phenotypes in a healthy ageing population. PLoS Genet. 8, e1002629. - PMC - PubMed
- Christensen K, Holm NV, McGue M, Corder L, Vaupel JW (1999) A Danish population‐based twin study on general health in the elderly. J. Aging Health 11, 49–64. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical