The Resource Identification Initiative: A cultural shift in publishing - PubMed (original) (raw)
doi: 10.12688/f1000research.6555.2. eCollection 2015.
Matthew Brush 2, Jeffery S Grethe 1, Melissa A Haendel 2, David N Kennedy 3, Sean Hill 4, Patrick R Hof 5, Maryann E Martone 1, Maaike Pols 6, Serena Tan 7, Nicole Washington 8, Elena Zudilova-Seinstra 9, Nicole Vasilevsky 2; Resource Identification Initiative Members are listed here: https://www.force11.org/node/4463/members
Affiliations
- PMID: 26594330
- PMCID: PMC4648211
- DOI: 10.12688/f1000research.6555.2
The Resource Identification Initiative: A cultural shift in publishing
Anita Bandrowski et al. F1000Res. 2015.
Abstract
A central tenet in support of research reproducibility is the ability to uniquely identify research resources, i.e., reagents, tools, and materials that are used to perform experiments. However, current reporting practices for research resources are insufficient to allow humans and algorithms to identify the exact resources that are reported or answer basic questions such as "What other studies used resource X?" To address this issue, the Resource Identification Initiative was launched as a pilot project to improve the reporting standards for research resources in the methods sections of papers and thereby improve identifiability and reproducibility. The pilot engaged over 25 biomedical journal editors from most major publishers, as well as scientists and funding officials. Authors were asked to include Research Resource Identifiers (RRIDs) in their manuscripts prior to publication for three resource types: antibodies, model organisms, and tools (including software and databases). RRIDs represent accession numbers assigned by an authoritative database, e.g., the model organism databases, for each type of resource. To make it easier for authors to obtain RRIDs, resources were aggregated from the appropriate databases and their RRIDs made available in a central web portal ( www.scicrunch.org/resources). RRIDs meet three key criteria: they are machine readable, free to generate and access, and are consistent across publishers and journals. The pilot was launched in February of 2014 and over 300 papers have appeared that report RRIDs. The number of journals participating has expanded from the original 25 to more than 40. Here, we present an overview of the pilot project and its outcomes to date. We show that authors are generally accurate in performing the task of identifying resources and supportive of the goals of the project. We also show that identifiability of the resources pre- and post-pilot showed a dramatic improvement for all three resource types, suggesting that the project has had a significant impact on reproducibility relating to research resources.
Keywords: Multi-centre initiative; Post-pilot data; Pre-pilot data; Publishing; Resource identifiers.
Conflict of interest statement
Competing interests: The authors declared no competing interests.
Figures
Figure 1.. The Resource Identification Initiative portal containing citable Research Resource Identifiers (RRIDs).
The workflow for authors is to visit
http://scicrunch.org/resources
, then select their resource type (see community resources box), type in search terms (note that the system attempts to expand known synonyms to improve search results) and open the “Cite This” dialog box. The dialog shown here displays the Invitrogen catalog number 80021 antibody with the RRID:AB_86329. The authors are asked to copy and paste this text into their methods section.
Figure 2.. RRIDs found in the published literature.
A. Google Scholar result for the anti-tyrosine hydroxylase antibody RRID (9/2014;
http://scholar.google.com/scholar?q=RRID:AB\_90755
).B. The most frequently reported RRIDs in the first 100 papers, by number of papers using the identifier. All data is available in the Supplementary Table and all identifiers can be accessed in Google Scholar.
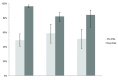
Figure 3.. Percent correctly reported RRIDs.
The percentage of resources that reported an RRID that pointed to the correct resource and with the correct syntax for each resource type is shown. The total number of resources for each type during the post-pilot is: primary antibodies, n = 429; organisms, n = 55; non-commercial tools, n = 78.
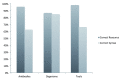
Figure 4.. Pre and post-pilot identifiability.
Resources (primary antibodies, organisms, and tools) were considered identifiable if they contained an accurate RRID or by using the same criteria as described in ( Vasilevsky_et al._, 2013). For tools (software and databases, which were not previously analyzed), these resources were considered identifiable if they contained an RRID or reported the manufacturer and version number. The total number of resources for each type is: primary antibodies pre-pilot, n = 140; primary antibodies post-pilot, n = 465; organisms pre-pilot, n = 58; organisms post-pilot, n = 139; non-commercial tools pre-pilot, n = 59; non-commercial tools post-pilot, n = 101. The y-axis is the average percent identifiable for each resource type. Variation from this average is shown by the bars: error bars indicate upper and lower 95% confidence intervals. Asterisks indicate significant difference by a z-score greater than 1.96.
Figure 5.. An exemplar third-party application using the RRID resolving service.
The “Antibody data for this article” application developed by Elsevier enhances articles on ScienceDirect. The application is available in 211 articles in 19 journals (more information can be found at:
http://www.elsevier.com/about/content-innovation/antibodies
).
Similar articles
- The Resource Identification Initiative: a cultural shift in publishing.
Bandrowski A, Brush M, Grethe JS, Haendel MA, Kennedy DN, Hill S, Hof PR, Martone ME, Pols M, Tan SC, Washington N, Zudilova-Seinstra E, Vasilevsky N; RINL Resource Identification Initiative. Bandrowski A, et al. Brain Behav. 2015 Dec 8;6(1):e00417. doi: 10.1002/brb3.417. eCollection 2016 Jan. Brain Behav. 2015. PMID: 27110440 Free PMC article. - The Resource Identification Initiative: A Cultural Shift in Publishing.
Bandrowski A, Brush M, Grethe JS, Haendel MA, Kennedy DN, Hill S, Hof PR, Martone ME, Pols M, Tan SS, Washington N, Zudilova-Seinstra E, Vasilevsky N; RINL Resource Identification Initiative. Bandrowski A, et al. Neuroinformatics. 2016 Apr;14(2):169-82. doi: 10.1007/s12021-015-9284-3. Neuroinformatics. 2016. PMID: 26589523 Free PMC article. - The Resource Identification Initiative: A Cultural Shift in Publishing.
Bandrowski A, Brush M, Grethe JS, Haendel MA, Kennedy DN, Hill S, Hof PR, Martone ME, Pols M, Tan SC, Washington N, Zudilova-Seinstra E, Vasilevsky N. Bandrowski A, et al. J Comp Neurol. 2016 Jan 1;524(1):8-22. doi: 10.1002/cne.23913. J Comp Neurol. 2016. PMID: 26599696 Free PMC article. - RRIDs: A Simple Step toward Improving Reproducibility through Rigor and Transparency of Experimental Methods.
Bandrowski AE, Martone ME. Bandrowski AE, et al. Neuron. 2016 May 4;90(3):434-6. doi: 10.1016/j.neuron.2016.04.030. Neuron. 2016. PMID: 27151636 Free PMC article. Review. - Alternatives to animal experimentation in basic research.
Gruber FP, Hartung T. Gruber FP, et al. ALTEX. 2004;21 Suppl 1:3-31. ALTEX. 2004. PMID: 15586255 Review.
Cited by
- Replacing bar graphs of continuous data with more informative graphics: are we making progress?
Riedel N, Schulz R, Kazezian V, Weissgerber T. Riedel N, et al. Clin Sci (Lond). 2022 Aug 12;136(15):1139-1156. doi: 10.1042/CS20220287. Clin Sci (Lond). 2022. PMID: 35822444 Free PMC article. - The landscape of nutri-informatics: a review of current resources and challenges for integrative nutrition research.
Chan L, Vasilevsky N, Thessen A, McMurry J, Haendel M. Chan L, et al. Database (Oxford). 2021 Jan 25;2021:baab003. doi: 10.1093/database/baab003. Database (Oxford). 2021. PMID: 33494105 Free PMC article. Review. - A High-Throughput Search for SFXN1 Physical Partners Led to the Identification of ATAD3, HSD10 and TIM50.
Tifoun N, Bekhouche M, De Las Heras JM, Guillaume A, Bouleau S, Guénal I, Mignotte B, Le Floch N. Tifoun N, et al. Biology (Basel). 2022 Aug 31;11(9):1298. doi: 10.3390/biology11091298. Biology (Basel). 2022. PMID: 36138777 Free PMC article. - How to avoid pitfalls in antibody use.
Pauly D, Hanack K. Pauly D, et al. F1000Res. 2015 Sep 7;4:691. doi: 10.12688/f1000research.6894.1. eCollection 2015. F1000Res. 2015. PMID: 26834988 Free PMC article. - Do organisms need an impact factor? Citations of key biological resources including model organisms reveal usage patterns and impact.
Piekniewska A, Anderson N, Roelandse M, Lloyd KCK, Korf I, Voss SR, de Castro G, Magnani DM, Varga Z, James-Zorn C, Horb M, Grethe JS, Bandrowski A. Piekniewska A, et al. bioRxiv [Preprint]. 2024 Jan 16:2024.01.15.575636. doi: 10.1101/2024.01.15.575636. bioRxiv. 2024. PMID: 38293091 Free PMC article. Preprint.
References
Grants and funding
- HHSN271200577531C/DA/NIDA NIH HHS/United States
- R24 OD011883/OD/NIH HHS/United States
- U24 DA039832/DA/NIDA NIH HHS/United States
- U24 DK097771/DK/NIDDK NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous