Emergence of Antimicrobial-Resistant Escherichia coli of Animal Origin Spreading in Humans - PubMed (original) (raw)
. 2016 Apr;33(4):898-914.
doi: 10.1093/molbev/msv280. Epub 2015 Nov 26.
Olivier Clermont 2, Thomas Guillard 3, Adrien Launay 2, Olga Danilchanka 4, Stéphanie Pons 3, Laure Diancourt 5, François Lebreton 4, Kristina Kadlec 6, Damien Roux 7, Deming Jiang 3, Sara Dion 2, Hugues Aschard 8, Maurice Denamur 2, Colette Cywes-Bentley 3, Stefan Schwarz 6, Olivier Tenaillon 2, Antoine Andremont 9, Bertrand Picard 10, John Mekalanos 4, Sylvain Brisse 5, Erick Denamur 11
Affiliations
- PMID: 26613786
- PMCID: PMC5013867
- DOI: 10.1093/molbev/msv280
Emergence of Antimicrobial-Resistant Escherichia coli of Animal Origin Spreading in Humans
David Skurnik et al. Mol Biol Evol. 2016 Apr.
Abstract
In the context of the great concern about the impact of human activities on the environment, we studied 403 commensal Escherichia coli/Escherichia clade strains isolated from several animal and human populations that have variable contacts to one another. Multilocus sequence typing (MLST) showed a decrease of diversity 1) in strains isolated from animals that had an increasing contact with humans and 2) in all strains that had increased antimicrobial resistance. A specific B1 phylogroup clonal complex (CC87, Institut Pasteur schema nomenclature) of animal origin was identified and characterized as being responsible for the increased antimicrobial resistance prevalence observed in strains from the environments with a high human-mediated antimicrobial pressure. CC87 strains have a high capacity of acquiring and disseminating resistance genes with specific metabolic and genetic determinants as demonstrated by high-throughput sequencing and phenotyping. They are good mouse gut colonizers but are not virulent. Our data confirm the predominant role of human activities in the emergence of antimicrobial resistance in the environmental bacterial strains and unveil a particular E. coli clonal complex of animal origin capable of spreading antimicrobial resistance to other members of microbial communities.
Keywords: Escherichia coli; antimicrobial resistance; clonal complex 87; commensal; phylogroup B1.
© The Author(s) 2015. Published by Oxford University Press on behalf of the Society for Molecular Biology and Evolution. All rights reserved. For permissions, please e-mail: journals.permissions@oup.com.
Figures
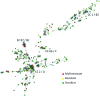
Fig. 1.
Analysis of the microevolutionary relationships between the 403 E. coli/Escherichia clade strains according to their MLST allelic profile using a minimum spanning tree algorithm. A total of 262 STs and 27 CCs encompassing 147 strains were identified. Among these 27 CCs, the 4 represented by more than 10 strains are indicated. They correspond to CC2 (phylogroup A, STc10 according to Achtman schema) (Wirth et al. 2006) (
http://mlst.warwick.ac.uk/mlst/dbs/Ecol
last accessed November 12, 2015), CC87 (phylogroup B1, STs 58 and 155 according to Achtman schema), CC66 (complete phylogroup C, STc23 according to Achtman schema), and CC1 (phylogroup B2, STc95 according to Achtman schema). The strains are colored according to their antimicrobial resistance pattern: susceptible to all antimicrobial agents, resistant to 1–2 antimicrobial agents, and resistant to at least three antimicrobial agents.
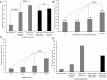
Fig. 2.
CC characteristics in the ROAR collection. Prevalence in percent of CCs in the 403 E. coli/Escherichia clade strains according to (A) animal and human vicinity and (B) antimicrobial resistance (absence = 0, 1, 2 or at least resistance to 3 antimicrobial agents). (C) Antimicrobial resistance expressed as RSs (Murray et al. 1990) in the main CCs (encompassing at least 10 strains) found in the studied population (CC1, CC2, CC66, and CC87). (D) Antimicrobial RSs according to the animal and human vicinity in CC87 strains. The star in the panel (D) indicates that no CC87 strain was found in the population of human without regular contact with animals.
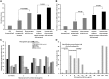
Fig. 3.
Antimicrobial resistance and CC87 in the ROAR collection. Antimicrobial RSs according to the animal and human vicinity in all the 403 E. coli/Escherichia clade strains of the collection (A), in all the strains of the collection excluding the CC87 (B), and in all the 403 strains of the collection according to the phylogenetic groups, the phylogroup B1 being considered with and without the CC87 strains (C). (D) Tetracycline MIC distribution of the E. coli/Escherichia clade strains detected as nonsusceptible by the disk diffusion method with the classification according to the CLSI clinical breakpoints. Note that only strains with a MIC > 64 µg/ml have a tet gene detected by multiplex PCR.
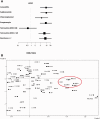
Fig. 4.
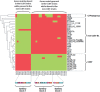
CC87 and statistical characteristics of the ROAR collection. (A) Odds ratio of antimicrobial resistance for strains from CC87. Resistance ≥ 2 corresponds resistance to at least two antimicrobial agents. (B) Factorial analysis of correspondence describing the associations among the various variables of the 403 E. coli/Escherichia clade strains. The variables in black are as follow: the phylogenetic groups (A, B1, B2, D, C, E, F, UG = ungrouped, clades = Escherichia clades), the VFs (kpsE, sfa, iroN, aer, pap, hly, cnf1, fyuA, afaD, stx1, stx2, eae and ST), the virulence categories (absence of VF = VF = 0, 1 to 3 VFs = VF = 1–3, 4 to 7 VFs = VF = 4–7), the resistance to seven antimicrobial agents (amoxicillin = AMX-R, sulphonamide = SUL-R, chloramphenicol = CHL-R, kanamycin = KAN-R, streptomycin = STR-R, nalidixic acid = NAL-R and tetracycline = TET-R), the detection of the tet genes, the MIC to tetracycline (MIC < 2 µg/ml, MIC = 2 to 16 µg/ml; MIC > 16 µg/ml), the effect of the efflux pump inhibitor PAβN (Phe-Arg-beta-naphtylamide) on the MIC of tetracycline, the three resistance categories (resistance to none of the antimicrobial agents tested = R = 0, resistance to 1 to 3 antimicrobial agents = R = 1–3, resistance to 4 to 7 antimicrobial agents = R = 4–7), the origins of the strains (wild animals = WA, domestic animals = DA, pet animals = PA, human with 2 populations: BIW and PF). The CCs are indicated in white. The association of CC87 with the plasmid-borne antimicrobial resistances is indicated by the circle.
Fig. 5.
CC87 and bacterial conjugation abilities. Efficiency of conjugation of (A) the β-lactamase gene _bla_TEM between two different donors (E. coli 14 and 38) and CC87 or non-CC87 strains, (B) the ESBL gene _bla_CTX-M-15 in direction of CC87 or non-CC87 strains, and (C) the same ESBL gene _bla_CTX-M-15 from CC87 strains to clinical and pathogenic strains. CC87 strains tested: ROAR61, 72, 82, 104 and 205. Non-CC87 strains tested: ROAR28, 79, 155, 171, 394.
Fig. 6.
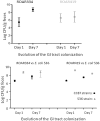
Commensal abilities of CC87 strains. Mouse digestive tract colonization assays. The day of inoculation, 104–105 E. coli ROAR bacteria were administered in 200 µl of PBS by oral route to mice free of coliform flora, either alone (A) or mixed at a ratio of 1:1 with the E. coli 536 strain, a well-known good colonizer of the mouse gut (Diard et al. 2010). On days 1 and 7 after bacterial administration, the sizes of bacterial populations in the intestine of mice were evaluated by plating dilutions of weighed fresh feces on LB agar with or without appropriate antimicrobial agents. Four mice were used per experiment. GI, gastrointestinal.
Fig. 7.
Specific genic presence/absence pattern of CC87 among B1 strains. The phylogeny of the B1 strains rooted on the phylogroup C was computed with 500 core genes. Gene presence/absence is illustrated for the set of genes that presented a contrasted signature between CC87 strains and non-CC87-B1. Among this set, four groups of linked genes are represented: metabolic genes; transporters; genes with alternative functions; and genes of unknown functions.
Fig. 8.
High-throughput Biolog phenotypic analysis of strains ROAR61 (CC87) and ROAR394 (non-CC87). (A) For each metabolite, the OD590nm after 24 h of growth is indicated. Metabolites influencing the bacterial growth of CC87 and non-CC87 are grouped according to the normalized ratios (ROAR61/ROAR394). Are represented: example of metabolites associated with an increased fitness of the non-CC87 strain (ratio < 0.5); example of metabolites with an increased fitness of the CC87 strain (ratio > 2); and no effect. (B) Individual growth curves of strains ROAR61 (square) and 394 (triangle) with (solid line) or without (dotted line) 1,10-phenanthroline (32 mg/l).
Fig. 9.
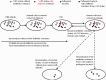
Schematic presentation of the acquisition and dissemination of antimicrobial resistance by CC87 strains.
Similar articles
- Virulence and antimicrobial resistance determinants of verotoxigenic Escherichia coli (VTEC) and of multidrug-resistant E. coli from foods of animal origin illegally imported to the EU by flight passengers.
Nagy B, Szmolka A, Smole Možina S, Kovač J, Strauss A, Schlager S, Beutlich J, Appel B, Lušicky M, Aprikian P, Pászti J, Tóth I, Kugler R, Wagner M. Nagy B, et al. Int J Food Microbiol. 2015 Sep 16;209:52-9. doi: 10.1016/j.ijfoodmicro.2015.06.026. Epub 2015 Jul 3. Int J Food Microbiol. 2015. PMID: 26148965 - High prevalence of ST131 among CTX-M-producing Escherichia coli from community-acquired infections, in the Czech Republic.
Papagiannitsis CC, Študentová V, Jakubů V, Španělová P, Urbášková P, Žemličková H, Hrabák J. Papagiannitsis CC, et al. Microb Drug Resist. 2015 Feb;21(1):74-84. doi: 10.1089/mdr.2014.0070. Epub 2014 Sep 4. Microb Drug Resist. 2015. PMID: 25188031 - The Role of Flies in Disseminating Plasmids with Antimicrobial-Resistance Genes Between Farms.
Usui M, Shirakawa T, Fukuda A, Tamura Y. Usui M, et al. Microb Drug Resist. 2015 Oct;21(5):562-9. doi: 10.1089/mdr.2015.0033. Epub 2015 May 20. Microb Drug Resist. 2015. PMID: 26061440 - Characterization of Pc promoter variants of class 1 integrons in Escherichia coli isolates from poultry meat.
Soufi L, Sáenz Y, Vinué L, Abbassi MS, Hammami S, Torres C. Soufi L, et al. Foodborne Pathog Dis. 2013 Dec;10(12):1075-7. doi: 10.1089/fpd.2013.1542. Epub 2013 Aug 29. Foodborne Pathog Dis. 2013. PMID: 23988017
Cited by
- Phenotypic and genotypic characterizations of bacteria isolated from the respiratory microbiota of healthy turkeys with potential for probiotic composition.
Trintinaglia M, de Brito KCT, Kobayashi RKT, Otutumi LK, Nakazato G, de Souza Gazal LE, Cruz VD, de Brito BG. Trintinaglia M, et al. Vet Res Commun. 2024 Feb;48(1):381-390. doi: 10.1007/s11259-023-10217-8. Epub 2023 Sep 14. Vet Res Commun. 2024. PMID: 37707656 - Highly Virulent and Multidrug-Resistant Escherichia coli Sequence Type 58 from a Sausage in Germany.
Eger E, Domke M, Heiden SE, Paditz M, Balau V, Huxdorff C, Zimmermann D, Homeier-Bachmann T, Schaufler K. Eger E, et al. Antibiotics (Basel). 2022 Jul 26;11(8):1006. doi: 10.3390/antibiotics11081006. Antibiotics (Basel). 2022. PMID: 35892394 Free PMC article. - A pilot study using environmental screening to determine the prevalence of Mycobacterium avium subspecies paratuberculosis (MAP) and antimicrobial resistance (AMR) in Irish cattle herds.
Ramovic E, Madigan G, McDonnell S, Griffin D, Bracken E, NiGhallchoir E, Quinless E, Galligan A, Egan J, Prendergast DM. Ramovic E, et al. Ir Vet J. 2020 Feb 15;73:3. doi: 10.1186/s13620-020-0156-2. eCollection 2020. Ir Vet J. 2020. PMID: 32082542 Free PMC article. - Occurrence and Molecular Characteristics of _Mcr-1_-Positive Escherichia coli from Healthy Meat Ducks in Shandong Province of China.
Liu F, Zhang R, Yang Y, Li H, Wang J, Lan J, Li P, Zhu Y, Xie Z, Jiang S. Liu F, et al. Animals (Basel). 2020 Jul 29;10(8):1299. doi: 10.3390/ani10081299. Animals (Basel). 2020. PMID: 32751361 Free PMC article. - Multidrug-Resistant and Clinically Relevant Gram-Negative Bacteria Are Present in German Surface Waters.
Falgenhauer L, Schwengers O, Schmiedel J, Baars C, Lambrecht O, Heß S, Berendonk TU, Falgenhauer J, Chakraborty T, Imirzalioglu C. Falgenhauer L, et al. Front Microbiol. 2019 Nov 29;10:2779. doi: 10.3389/fmicb.2019.02779. eCollection 2019. Front Microbiol. 2019. PMID: 31849911 Free PMC article.
References
- Agersø Y, Guardabassi L. 2005. Identification of Tet 39, a novel class of tetracycline resistance determinant in Acinetobacter spp. of environmental and clinical origin. J Antimicrob Chemother. 55:566–569. - PubMed
- Andersson DI, Hughes D. 2010. Antibiotic resistance and its cost: is it possible to reverse resistance? Nat Rev Microbiol. 8:260–271. - PubMed
- Andremont A. 2003. Commensal flora may play key role in spreading antibiotic resistance. ASM News 63:601–607.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources