Networks of energetic and metabolic interactions define dynamics in microbial communities - PubMed (original) (raw)
Networks of energetic and metabolic interactions define dynamics in microbial communities
Mallory Embree et al. Proc Natl Acad Sci U S A. 2015.
Abstract
Microorganisms form diverse communities that have a profound impact on the environment and human health. Recent technological advances have enabled elucidation of community diversity at high resolution. Investigation of microbial communities has revealed that they often contain multiple members with complementing and seemingly redundant metabolic capabilities. An understanding of the communal impacts of redundant metabolic capabilities is currently lacking; specifically, it is not known whether metabolic redundancy will foster competition or motivate cooperation. By investigating methanogenic populations, we identified the multidimensional interspecies interactions that define composition and dynamics within syntrophic communities that play a key role in the global carbon cycle. Species-specific genomes were extracted from metagenomic data using differential coverage binning. We used metabolic modeling leveraging metatranscriptomic information to reveal and quantify a complex intertwined system of syntrophic relationships. Our results show that amino acid auxotrophies create additional interdependencies that define community composition and control carbon and energy flux through the system while simultaneously contributing to overall community robustness. Strategic use of antimicrobials further reinforces this intricate interspecies network. Collectively, our study reveals the multidimensional interactions in syntrophic communities that promote high species richness and bolster community stability during environmental perturbations.
Keywords: interspecies interactions; metabolic modeling; methanogens; microbial communities; microbiome.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Fig. 1.
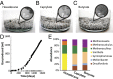
Growth of methanogenic communities. (A) Teflon boiling stones were coated with hexadecane to increase the contact between cells and the hydrophobic substrate. The methanogenic community propagated in 300 mL mineral medium (6) containing 20 mM hexadecane grows very slowly to low biomass density. The hexadecane community was passed onto medium containing (B) 2 mM caprylate or (C) 5 mM butyrate. All communities grew to very low biomass densities, despite being very metabolically active as indicated by gas (methane) bubbles formed. (D) The methanogenic community formed a total of 1,407 mL methane after 1,551 d of incubation (black circles). The control containing no substrate (white circles) did not form any methane. Growth curves of the fatty acid cultures were previously published (7). (E) Metagenomic sequencing reads were mapped back to the newly acquired genomes to establish species abundance in the community under hexadecane-, caprylate-, and butyrate-degrading conditions. Numbers of mapped reads were normalized by genome size.
Fig. S1.
Genomes of individual community members. Differential coverage metagenomic binning (9) enabled the extraction of seven genomes. Sequence homologies of genomes obtained were compared with their closest relatives. The Smithella sp. ME-1 genome was sourced from the same community by a single-cell approach (4), explaining the high sequence similarity.
Fig. 2.
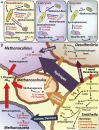
Complex interspecies interactions driving syntrophy. (A) Metabolic interspecies interactions in three communities as determined by transcriptomic information. (B) Interspecies exchanges as calculated by metabolic modeling using physiological data for the hexadecane uptake and methane production rates. Flux values are being reported as absolute values (extrapolated over the time represented by the physiological measurements) rather than rate. Hexadecane initially degraded by Smithella results in reducing equivalents that are passed to its syntrophic partners Methanoculleus, Methanocalculus, Methanosaeta, and Desulfovibrio in various forms. Desulfovibrio is converting formate to hydrogen and CO2, possibly forming its own syntrophic interactions with the methanogens.
Fig. 3.
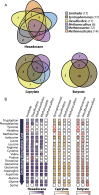
Amino acid auxotrophies shape community composition. (A) Overlap of amino acid biosynthesis capabilities between community members in communities growing with hexadecane, caprylate, or butyrate. Numbers in parentheses refer to the numbers of amino acids that the particular species can produce. (B) Specific amino acid auxotrophies present in each species for each community. Amino acids have been ranked according to biosynthetic cost (arrow) (8). A colored square denotes that a species can synthesize an amino acid. The intensity of each color (based on the scale) represents the relative expression of the synthesis pathways (Methods), with darker/more intense color indicating higher expression. Amino acids exclusively produced by one microorganism are highlighted in red.
Similar articles
- Genome-centric resolution of microbial diversity, metabolism and interactions in anaerobic digestion.
Vanwonterghem I, Jensen PD, Rabaey K, Tyson GW. Vanwonterghem I, et al. Environ Microbiol. 2016 Sep;18(9):3144-58. doi: 10.1111/1462-2920.13382. Epub 2016 Jul 22. Environ Microbiol. 2016. PMID: 27317862 - Ecological and evolutionary interactions in syntrophic methanogenic consortia.
Kato S, Watanabe K. Kato S, et al. Microbes Environ. 2010;25(3):145-51. doi: 10.1264/jsme2.me10122. Microbes Environ. 2010. PMID: 21576866 Review. - Metabolic dependencies drive species co-occurrence in diverse microbial communities.
Zelezniak A, Andrejev S, Ponomarova O, Mende DR, Bork P, Patil KR. Zelezniak A, et al. Proc Natl Acad Sci U S A. 2015 May 19;112(20):6449-54. doi: 10.1073/pnas.1421834112. Epub 2015 May 4. Proc Natl Acad Sci U S A. 2015. PMID: 25941371 Free PMC article. - Community Structure in Methanogenic Enrichments Provides Insight into Syntrophic Interactions in Hydrocarbon-Impacted Environments.
Fowler SJ, Toth CR, Gieg LM. Fowler SJ, et al. Front Microbiol. 2016 Apr 22;7:562. doi: 10.3389/fmicb.2016.00562. eCollection 2016. Front Microbiol. 2016. PMID: 27148240 Free PMC article. - Network-based metabolic analysis and microbial community modeling.
Cardona C, Weisenhorn P, Henry C, Gilbert JA. Cardona C, et al. Curr Opin Microbiol. 2016 Jun;31:124-131. doi: 10.1016/j.mib.2016.03.008. Epub 2016 Apr 6. Curr Opin Microbiol. 2016. PMID: 27060776 Review.
Cited by
- Molecular Microbial Community Analysis as an Analysis Tool for Optimal Biogas Production.
Hashemi S, Hashemi SE, Lien KM, Lamb JJ. Hashemi S, et al. Microorganisms. 2021 May 28;9(6):1162. doi: 10.3390/microorganisms9061162. Microorganisms. 2021. PMID: 34071282 Free PMC article. Review. - Metabolic exchanges are ubiquitous in natural microbial communities.
Kost C, Patil KR, Friedman J, Garcia SL, Ralser M. Kost C, et al. Nat Microbiol. 2023 Dec;8(12):2244-2252. doi: 10.1038/s41564-023-01511-x. Epub 2023 Nov 23. Nat Microbiol. 2023. PMID: 37996708 Review. - Engineered Interspecies Amino Acid Cross-Feeding Increases Population Evenness in a Synthetic Bacterial Consortium.
Ziesack M, Gibson T, Oliver JKW, Shumaker AM, Hsu BB, Riglar DT, Giessen TW, DiBenedetto NV, Bry L, Way JC, Silver PA, Gerber GK. Ziesack M, et al. mSystems. 2019 Aug 13;4(4):e00352-19. doi: 10.1128/mSystems.00352-19. mSystems. 2019. PMID: 31409662 Free PMC article. - Metatranscriptomics-guided genome-scale metabolic modeling of microbial communities.
Zampieri G, Campanaro S, Angione C, Treu L. Zampieri G, et al. Cell Rep Methods. 2023 Jan 6;3(1):100383. doi: 10.1016/j.crmeth.2022.100383. eCollection 2023 Jan 23. Cell Rep Methods. 2023. PMID: 36814842 Free PMC article. - Comparative Genomics of Nitrogen Cycling Pathways in Bacteria and Archaea.
Albright MBN, Timalsina B, Martiny JBH, Dunbar J. Albright MBN, et al. Microb Ecol. 2019 Apr;77(3):597-606. doi: 10.1007/s00248-018-1239-4. Epub 2018 Aug 13. Microb Ecol. 2019. PMID: 30105504
References
- Gans J, Wolinsky M, Dunbar J. Computational improvements reveal great bacterial diversity and high metal toxicity in soil. Science. 2005;309(5739):1387–1390. - PubMed
- Martin W, Müller M. The hydrogen hypothesis for the first eukaryote. Nature. 1998;392(6671):37–41. - PubMed
- Dolfing J, Larter SR, Head IM. Thermodynamic constraints on methanogenic crude oil biodegradation. ISME J. 2008;2(4):442–452. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases