AIM2 inflammasome in infection, cancer, and autoimmunity: Role in DNA sensing, inflammation, and innate immunity - PubMed (original) (raw)
Review
AIM2 inflammasome in infection, cancer, and autoimmunity: Role in DNA sensing, inflammation, and innate immunity
Si Ming Man et al. Eur J Immunol. 2016 Feb.
Abstract
Recognition of DNA by the cell is an important immunological signature that marks the initiation of an innate immune response. AIM2 is a cytoplasmic sensor that recognizes dsDNA of microbial or host origin. Upon binding to DNA, AIM2 assembles a multiprotein complex called the inflammasome, which drives pyroptosis and proteolytic cleavage of the proinflammatory cytokines pro-IL-1β and pro-IL-18. Release of microbial DNA into the cytoplasm during infection by Francisella, Listeria, Mycobacterium, mouse cytomegalovirus, vaccinia virus, Aspergillus, and Plasmodium species leads to activation of the AIM2 inflammasome. In contrast, inappropriate recognition of cytoplasmic self-DNA by AIM2 contributes to the development of psoriasis, dermatitis, arthritis, and other autoimmune and inflammatory diseases. Inflammasome-independent functions of AIM2 have also been described, including the regulation of the intestinal stem cell proliferation and the gut microbiota ecology in the control of colorectal cancer. In this review we provide an overview of the latest research on AIM2 inflammasome and its role in infection, cancer, and autoimmunity.
Keywords: AIM2 inflammasome; Autoimmunity; Bacterial/viral infection; Cancer; DNA sensing; Gut microbiota.
© 2015 WILEY-VCH Verlag GmbH & Co. KGaA, Weinheim.
Conflict of interest statement
Conflict of interest
The authors declare no financial or commercial conflict of interest.
Figures
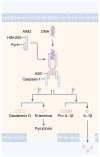
Figure 1. The molecular basis for the activation of the AIM2 inflammasome
The DNA sensor AIM2 is composed of an N-terminal pyrin domain and a C-terminal HIN-200 domain. The pyrin and HIN-200 domain of AIM2 form an intramolecular complex and are maintained in an autoinhibitory state. Cytoplasmic dsDNA induces activation of AIM2. The HIN-200 domain interacts with dsDNA in a sequence-independent manner, by binding to the sugar-phosphate backbone of dsDNA. The pyrin domain of AIM2 binds to the pyrin domain of ASC. CARD of ASC binds the CARD of pro-caspase-1, forming a macromolecular complex known as the AIM2 inflammasome. Activated caspase-1 drives cleavage of pro-IL-1β and pro-IL-18. Caspase-1 also cleaves the substrate gasdermin D. The N-terminal fragment of gasdermin D induces pyroptosis, allowing mature IL-1β and IL-18 to be released from the cell.
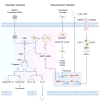
Figure 2. Regulation of the activation of the AIM2 inflammasome
The AIM2 inflammasome is activated by a number of microbial pathogens and dsDNA ligands, including the DNA virus mouse cytomegalovirus (MCMV), the cytosolic bacterium Francisella novicida and the dsDNA ligand poly(dA:dT). MCMV infection or transfection of poly(dA:dT) leads to “canonical” activation of the AIM2 inflammasome, which does not require the type I interferon (IFN) pathway. F. novicida infection activates the AIM2 inflammasome via a “non-canonical” pathway owing to its requirement for type I IFN, analogous to the non-canonical NLRP3 inflammasome pathway. Intracellular F. novicida releases DNA into the cytoplasm to activate the DNA sensors cGAS, STING and IFI204, which drive transcription of genes encoding type I IFN molecules. It remains unclear why the released DNA is unable to activate AIM2 at this stage, since AIM2 is constitutively expressed in the cell. Type I IFN provides a feedback loop to induce expression of the transcription factor IRF1, which upregulates expression of the IFN-inducible GTPases, including GBP2 and GBP5. GBP2 and GBP5 are recruited to bacterial structures, however, whether they directly target the bacterial membrane or the membrane of intact _Francisella_-containing vacuole is unclear. Nevertheless, GBPs mediate bacterial killing, resulting in abundant release of bacterial DNA for recognition by AIM2. Assembly of the AIM2 inflammasome induces caspase-1-dependent cleavage of pro-IL-1β and pro-IL-18. Caspase-1 also drives cleavage of the substrate gasdermin D to induce pyroptosis.
Similar articles
- Role of AIM2 inflammasome in inflammatory diseases, cancer and infection.
Sharma BR, Karki R, Kanneganti TD. Sharma BR, et al. Eur J Immunol. 2019 Nov;49(11):1998-2011. doi: 10.1002/eji.201848070. Epub 2019 Aug 14. Eur J Immunol. 2019. PMID: 31372985 Free PMC article. Review. - The AIM2 inflammasome: Sensor of pathogens and cellular perturbations.
Lugrin J, Martinon F. Lugrin J, et al. Immunol Rev. 2018 Jan;281(1):99-114. doi: 10.1111/imr.12618. Immunol Rev. 2018. PMID: 29247998 Review. - Regulation of inflammasome activation.
Man SM, Kanneganti TD. Man SM, et al. Immunol Rev. 2015 May;265(1):6-21. doi: 10.1111/imr.12296. Immunol Rev. 2015. PMID: 25879280 Free PMC article. Review. - AIM2 in health and disease: Inflammasome and beyond.
Kumari P, Russo AJ, Shivcharan S, Rathinam VA. Kumari P, et al. Immunol Rev. 2020 Sep;297(1):83-95. doi: 10.1111/imr.12903. Epub 2020 Jul 26. Immunol Rev. 2020. PMID: 32713036 Free PMC article. Review. - Human hepatocytes express absent in melanoma 2 and respond to hepatitis B virus with interleukin-18 expression.
Pan X, Xu H, Zheng C, Li M, Zou X, Cao H, Xu Q. Pan X, et al. Virus Genes. 2016 Aug;52(4):445-52. doi: 10.1007/s11262-016-1327-9. Epub 2016 Apr 19. Virus Genes. 2016. PMID: 27094165
Cited by
- The Role of the AIM2 Gene in Obesity-Related Glucose and Lipid Metabolic Disorders: A Recent Update.
Zhang Y, Xuan X, Ye D, Liu D, Song Y, Gao F, Lu S. Zhang Y, et al. Diabetes Metab Syndr Obes. 2024 Oct 21;17:3903-3916. doi: 10.2147/DMSO.S488978. eCollection 2024. Diabetes Metab Syndr Obes. 2024. PMID: 39465122 Free PMC article. Review. - Cytoplasmic DNA and AIM2 inflammasome in RA: where they come from and where they go?
Xu C, Jing W, Liu C, Yuan B, Zhang X, Liu L, Zhang F, Chen P, Liu Q, Wang H, Du X. Xu C, et al. Front Immunol. 2024 Oct 10;15:1343325. doi: 10.3389/fimmu.2024.1343325. eCollection 2024. Front Immunol. 2024. PMID: 39450183 Free PMC article. Review. - Inhibition of NLRP3 inflammasome contributes to paclitaxel efficacy in triple negative breast cancer treatment.
Balahura Stămat LR, Dinescu S. Balahura Stămat LR, et al. Sci Rep. 2024 Oct 21;14(1):24753. doi: 10.1038/s41598-024-75805-3. Sci Rep. 2024. PMID: 39433537 Free PMC article. - Applications of pyroptosis activators in tumor immunotherapy.
Bao X, Sun M, Meng L, Zhang H, Yi X, Zhang P. Bao X, et al. Mater Today Bio. 2024 Aug 6;28:101191. doi: 10.1016/j.mtbio.2024.101191. eCollection 2024 Oct. Mater Today Bio. 2024. PMID: 39221221 Free PMC article. Review. - Research progress on the effect of pyroptosis on the occurrence, development, invasion and metastasis of colorectal cancer.
Wang X, Yin QH, Wan LL, Sun RL, Wang G, Gu JF, Tang DC. Wang X, et al. World J Gastrointest Oncol. 2024 Aug 15;16(8):3410-3427. doi: 10.4251/wjgo.v16.i8.3410. World J Gastrointest Oncol. 2024. PMID: 39171180 Free PMC article. Review.
References
- Hemmi H, Takeuchi O, Kawai T, Kaisho T, Sato S, Sanjo H, Matsumoto M, Hoshino K, Wagner H, Takeda K, Akira S. A Toll-like receptor recognizes bacterial DNA. Nature. 2000;408:740–745. - PubMed
- Ishii KJ, Coban C, Kato H, Takahashi K, Torii Y, Takeshita F, Ludwig H, Sutter G, Suzuki K, Hemmi H, Sato S, Yamamoto M, Uematsu S, Kawai T, Takeuchi O, Akira S. A Toll-like receptor-independent antiviral response induced by double-stranded B-form DNA. Nat Immunol. 2006;7:40–48. - PubMed
- Stetson DB, Medzhitov R. Recognition of cytosolic DNA activates an IRF3-dependent innate immune response. Immunity. 2006;24:93–103. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- CA163507/CA/NCI NIH HHS/United States
- R01 AI101935/AI/NIAID NIH HHS/United States
- R37 AI101935/AI/NIAID NIH HHS/United States
- AI101935/AI/NIAID NIH HHS/United States
- R01 AR056296/AR/NIAMS NIH HHS/United States
- AR056296/AR/NIAMS NIH HHS/United States
- R01 CA163507/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources