A Role of TDIF Peptide Signaling in Vascular Cell Differentiation is Conserved Among Euphyllophytes - PubMed (original) (raw)
A Role of TDIF Peptide Signaling in Vascular Cell Differentiation is Conserved Among Euphyllophytes
Yuki Hirakawa et al. Front Plant Sci. 2015.
Abstract
Peptide signals mediate a variety of cell-to-cell communication crucial for plant growth and development. During Arabidopsis thaliana vascular development, a CLE (CLAVATA3/EMBRYO SURROUNDING REGION-related) family peptide hormone, TDIF (tracheary element differentiation inhibitory factor), regulates procambial cell fate by its inhibitory activity on xylem differentiation. To address if this activity is conserved among vascular plants, we performed comparative analyses of TDIF signaling in non-flowering vascular plants (gymnosperms, ferns and lycophytes). We identified orthologs of TDIF/CLE as well as its receptor TDR/PXY (TDIF RECEPTOR/PHLOEM INTERCALATED WITH XYLEM) in Ginkgo biloba, Adiantum aethiopicum, and Selaginella kraussiana by RACE-PCR. The predicted TDIF peptide sequences in seed plants and ferns were identical to that of A. thaliana TDIF. We examined the effects of exogenous CLE peptide-motif sequences of TDIF in these species. We found that liquid culturing of dissected leaves or shoots was useful for examining TDIF activity during vascular development. TDIF treatment suppressed xylem/tracheary element differentiation of procambial cells in G. biloba and A. aethiopicum leaves. In contrast, neither TDIF nor putative endogenous TDIF inhibited xylem differentiation in developing shoots and rhizophores of S. kraussiana. These data suggest that activity of TDIF in vascular development is conserved among extant euphyllophytes. In addition to the conserved function, via liquid culturing of its bulbils, we found a novel inhibitory activity on root growth in the fern Asplenium × lucrosum suggesting lineage-specific co-option of peptide signaling occurred during the evolution of vascular plant organs.
Keywords: CLE peptides; LRR-RLKs; non-model organism; plant evo-devo; plant vascular development; vascular plants.
Figures
Figure 1
TDIF/H-type CLE genes in land plants and the bioactivities of CLE peptides in Arabidopsis thaliana. (A) Alignment of deduced primary sequences for TDIF genes identified in this study (Ginkgo biloba CLE1, Adiantum aethiopicum CLE1, Selaginella kraussiana CLE1) with A. thaliana CLE41 and Selaginella moellendorffii CLE14. Gray and blue texts indicate signal peptide and the 12 amino-acid CLE peptide motifs, respectively. (B–E) Effects of peptides in A. thaliana plants grown for 10 days in liquid medium containing no additional peptide (B), 5 μM TDIF (C), 5 μM SkCLE1 peptide (D), or 5 μM SkCLE1L peptide (E). Yellow arrows indicate veins without visible xylem vessels. Scale bars: 100 μm.
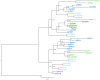
Figure 2
Phylogenetic relationships of vascular plant TDR genes. The phylogram was generated with the kinase domain sequences based on a Bayesian method. The posterior probabilities of trees are shown at the nodes. Using AtER as an outgroup, CLV and TDR genes each form monophyletic clades. Genes are color coded by taxon: moss, purple; lycophyte, light blue; ferns, dark blue; gymnosperm, dark green; angiosperm, light green. Species abbreviations are as follows: Pp, Physcomitrella patens; Sk, Selaginella kraussiana; Sm, Selaginella moellendorfii; Aa, Adiantum aethiopicum; Af, Azolla filiculoides; Eg, Equisetum giganteum; Paq, Pteridium aquilinum; Gb, Ginkgo biloba; Pab, Picea abies; Pg, Picea glauca; At, Arabidopsis thaliana. The paired S. moellendorfii sequences (A and B) are likely alleles.
Figure 3
Effects of TDIF treatment in Ginkgo biloba leaf. (A–H) Veins of G. biloba leaves cultured for 10 days in liquid medium containing no additional peptide (A,C,E,G) or 10 μM TDIF (B,D,F,H). (A–F) Images near the edge (A,B) or middle (C–F) of the leaf blade. Yellow arrows indicate veins without visible tracheids. Mucilage canals are indicated as mc. (G,H) Cross section of the veins. Black and red arrowheads indicate phloem and xylem, respectively. (I) Overall leaf morphology before (left) and after (right) 10-day culture. The dashed line illustrates the approximate position of sectioning plane for (G,H). Scale bars: 500 μm in (A–D), 100 μm in (E,F), 50 μm in (G,H), and 0.5 cm in (I).
Figure 4
Effects of TDIF treatment in fern fronds. Adiantum aethiopicum (A,B) and Asplenium × lucrosum (C,D) fronds from plants cultured for 10 or 51 days in liquid medium containing no additional peptide (A,C) or 10 μM TDIF (B,D). Yellow arrows indicate discontinued xylem strand in (B) and veins without visible tracheids in (D). Scale bars: 200 μm.
Figure 5
Effects of TDIF on the morphology of Asplenium × lucrosum. (A–D) Overall morphology of A.× lucrosum plants grown for 3 months in liquid medium containing no additional peptide (A), 100 nM TDIF (B), 1 μM TDIF (C), or 10 μM TDIF (D). (E) comparison of root morphology grown for 5 weeks in liquid culture containing different concentration of TDIF peptides as indicated. (F–I) Cross sections at the middle of the roots grown in control (F,H) or 1 μM TDIF (G,I) medium. Approximate positions for sectioning were illustrated in (E) by arrowheads. The images for (H,I) are magnification of central cylinder in (F,G). Arrows in (H) indicate protoxylem poles. Scale bars: 2 cm in (A–D), 1 cm in (E), 100 μm in (F,G), and 50 μm in (H,I).
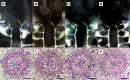
Figure 6
Effects of TDIF in Selaginella kraussiana. Cleared whole-mount images of shoot meristem region with xylem strands (A–D) and cross sections of rhizophores (E–H) of S. kraussiana shoots grown for 3 weeks in liquid medium containing no additional peptide (A,E), 5 μM TDIF (B,F), 5 μM SkCLE1 peptide (C,G), or 5 μM SKCLE1L peptide (D,H). Yellow arrows in (A–D) indicate the termini of xylem strands (white lines in images) just below the shoot apical meristem. Scale bars: 100 μm (A–D), 20 μm (E–H).
Similar articles
- Crystal structure of PXY-TDIF complex reveals a conserved recognition mechanism among CLE peptide-receptor pairs.
Zhang H, Lin X, Han Z, Qu LJ, Chai J. Zhang H, et al. Cell Res. 2016 May;26(5):543-55. doi: 10.1038/cr.2016.45. Epub 2016 Apr 8. Cell Res. 2016. PMID: 27055373 Free PMC article. - Regulation of vascular development by CLE peptide-receptor systems.
Hirakawa Y, Kondo Y, Fukuda H. Hirakawa Y, et al. J Integr Plant Biol. 2010 Jan;52(1):8-16. doi: 10.1111/j.1744-7909.2010.00904.x. J Integr Plant Biol. 2010. PMID: 20074136 Review. - A membrane-associated NAC domain transcription factor XVP interacts with TDIF co-receptor and regulates vascular meristem activity.
Yang JH, Lee KH, Du Q, Yang S, Yuan B, Qi L, Wang H. Yang JH, et al. New Phytol. 2020 Apr;226(1):59-74. doi: 10.1111/nph.16289. Epub 2019 Dec 5. New Phytol. 2020. PMID: 31660587 - Non-cell-autonomous control of vascular stem cell fate by a CLE peptide/receptor system.
Hirakawa Y, Shinohara H, Kondo Y, Inoue A, Nakanomyo I, Ogawa M, Sawa S, Ohashi-Ito K, Matsubayashi Y, Fukuda H. Hirakawa Y, et al. Proc Natl Acad Sci U S A. 2008 Sep 30;105(39):15208-13. doi: 10.1073/pnas.0808444105. Epub 2008 Sep 23. Proc Natl Acad Sci U S A. 2008. PMID: 18812507 Free PMC article. - CLE peptides: critical regulators for stem cell maintenance in plants.
Song XF, Hou XL, Liu CM. Song XF, et al. Planta. 2021 Nov 29;255(1):5. doi: 10.1007/s00425-021-03791-1. Planta. 2021. PMID: 34841457 Review.
Cited by
- Functional Modules in the Meristems: "Tinkering" in Action.
Kuznetsova K, Efremova E, Dodueva I, Lebedeva M, Lutova L. Kuznetsova K, et al. Plants (Basel). 2023 Oct 23;12(20):3661. doi: 10.3390/plants12203661. Plants (Basel). 2023. PMID: 37896124 Free PMC article. Review. - The CLV3 Homolog in Setaria viridis Selectively Controls Inflorescence Meristem Size.
Zhu C, Liu L, Crowell O, Zhao H, Brutnell TP, Jackson D, Kellogg EA. Zhu C, et al. Front Plant Sci. 2021 Feb 15;12:636749. doi: 10.3389/fpls.2021.636749. eCollection 2021. Front Plant Sci. 2021. PMID: 33659018 Free PMC article. - MOL1 is required for cambium homeostasis in Arabidopsis.
Gursanscky NR, Jouannet V, Grünwald K, Sanchez P, Laaber-Schwarz M, Greb T. Gursanscky NR, et al. Plant J. 2016 May;86(3):210-20. doi: 10.1111/tpj.13169. Epub 2016 Apr 14. Plant J. 2016. PMID: 26991973 Free PMC article. - Cryptic bioactivity capacitated by synthetic hybrid plant peptides.
Hirakawa Y, Shinohara H, Welke K, Irle S, Matsubayashi Y, Torii KU, Uchida N. Hirakawa Y, et al. Nat Commun. 2017 Feb 6;8:14318. doi: 10.1038/ncomms14318. Nat Commun. 2017. PMID: 28165456 Free PMC article. - Peptide Signaling Pathways Regulate Plant Vascular Development.
Yuan B, Wang H. Yuan B, et al. Front Plant Sci. 2021 Sep 3;12:719606. doi: 10.3389/fpls.2021.719606. eCollection 2021. Front Plant Sci. 2021. PMID: 34539713 Free PMC article. Review.
References
- Araya T., Miyamoto M., Wibowo J., Suzuki A., Kojima S., Tsuchiya Y. N., et al. . (2014). CLE-CLAVATA1 peptide-receptor signaling module regulates the expansion of plant root systems in a nitrogen-dependent manner. Proc. Natl. Acad. Sci. U.S.A. 111, 2029–2034. 10.1073/pnas.1319953111 - DOI - PMC - PubMed
- Clark S. E., Running M. P., Meyerowitz E. M. (1995). CLAVATA3 is a specific regulator of shoot and floral meristem development affecting the same processes as CLAVATA1. Development 121, 2057–2067.
- Depuydt S., Rodriguez-Villalon A., Santuari L., Wyser-Rmili C., Ragni L., Hardtke C. S. (2013). Suppression of Arabidopsis protophloem differentiation and root meristem growth by CLE45 requires the receptor-like kinase BAM3. Proc. Natl. Acad. Sci. U.S.A. 110, 7074–7079. 10.1073/pnas.1222314110 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases