Circular RNAs are long-lived and display only minimal early alterations in response to a growth factor - PubMed (original) (raw)
Circular RNAs are long-lived and display only minimal early alterations in response to a growth factor
Yehoshua Enuka et al. Nucleic Acids Res. 2016.
Abstract
Circular RNAs (circRNAs) are widespread circles of non-coding RNAs with largely unknown function. Because stimulation of mammary cells with the epidermal growth factor (EGF) leads to dynamic changes in the abundance of coding and non-coding RNA molecules, and culminates in the acquisition of a robust migratory phenotype, this cellular model might disclose functions of circRNAs. Here we show that circRNAs of EGF-stimulated mammary cells are stably expressed, while mRNAs and microRNAs change within minutes. In general, the circRNAs we detected are relatively long-lived and weakly expressed. Interestingly, they are almost ubiquitously co-expressed with the corresponding linear transcripts, and the respective, shared promoter regions are more active compared to genes producing linear isoforms with no detectable circRNAs. These findings imply that altered abundance of circRNAs, unlike changes in the levels of other RNAs, might not play critical roles in signaling cascades and downstream transcriptional networks that rapidly commit cells to specific outcomes.
© The Author(s) 2015. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures
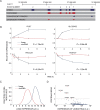
Figure 1.
Unlike mRNAs and microRNAs, circular RNAs of mammary cells display minor changes in response to an extracellular cue. (A) Venn diagrams presenting genomic origins of circRNAs found in MCF10A mammary cells. The analysis comprises 1498 circRNA molecules we identified in MCF10A cells using RNA-sequencing. Of these, 1451 molecules (97%) overlap known transcripts. The remainder 47 circRNAs are either intergenic (14 transcripts) or antisense to known transcripts (N = 33). Inner circles represent Quantified fractions, meaning transcripts we followed also by using PCR; For circRNAs derived from known transcripts, the majority (>90%) of the Quantified fraction refers to circRNAs, the response of which to EGF was assayed using divergent and convergent sets of primers, while for the remainder of the Quantified fraction, including circRNAs derived from either antisense or intergenic regions, measurements were performed using only divergent sets of primers. (B) MCF10A human mammary epithelial cells were starved overnight for serum factors. Thereafter they were treated with EGF (10 ng/ml) for the indicated time intervals. High-throughput PCR and specific primers were applied on isolated RNA samples to amplify 241 of 1498 circRNA species previously identified using RNA sequencing. This group included >50% of the most abundant candidates, along with dozens of other circRNAs with varying expression levels. The presented heatmap (right panel) depicts time-dependent alterations in expression levels of specific circRNAs. These alterations are compared to mRNA and microRNA alterations we previously observed, using microarrays, while stimulating MCF10A under identical conditions (5,36). Note that all previously analyzed miRNAs are represented, but in order to match the size of the circRNA population, only randomly selected, 16.1% of all MCF10A's mRNA molecules, are depicted in the heatmap. Data were normalized to time zero and ordered according to the time point corresponding to the maximal change. Red squares represent an increase and blue squares represent a decrease, as shown in the scale bar on the right. CircRNA results represent biological duplicates performed in technical triplicates. (C) A histogram showing the range of abundance changes of mRNAs, miRNAs and circRNAs (N = 3608, N = 164 and N = 288, respectively) displayed by EGF-stimulated MCF10A cells. To construct the histogram, the maximal change value (induction or repression) along the stimulation interval (240 min) was found for each RNA molecule. Note that circRNAs exhibit narrower dynamic range (highlighted region) than mRNAs and miRNAs (P < 1e-100, F-test, Bonferroni corrected for multiple comparisons). Note that only 3 time points (30, 90 and 240 minutes) were available for 47 of the presented circRNAs.
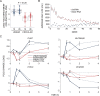
Figure 2.
circRNAs are less dynamic and abundant relative to linear isoforms derived from the same host gene. (A) A linear diagram exemplifying primer pairs (arrows) for parallel measurements of levels of circular and linear RNA isoforms derived from the same host gene. The ERBB2 (chromosome 17) gene is shown as an example. The respective circRNA (red) spans five internal exons. The divergent primers (red arrows; circular transcript) flank the non-canonical splicing (circularization), whereas the convergent primers (blue arrows) were designed for measuring the corresponding linear isoform. (B) PCR and four pairs of primers (designed as in A) were used to follow the response of the indicated linear and circular isoforms of four EGF-regulated genes. Note that mRNAs corresponding to PUS7 and SLC25A32 are induced when MCF10A cells are stimulated with EGF, whereas PCMTD1 and PIK3C2G mRNAs undergo down-regulation under the same conditions. Shown are means ± S.E. of triplicates. _P_-values were calculated using two-way Anova with time and RNA type as categorical factors. (C) A histogram comparing the abundance of circRNAs and the corresponding linear transcripts (N = 203) of MCF10A mammary cells. Note that circRNAs are, on average, 36 times less abundant than the corresponding linear isoforms (P = 5.45e-78, _t_-test, two-tail, paired; A.U., arbitrary units). (D) A dot plot presenting the ratio between expression levels of each circRNA of MCF10A cells we characterized and the respective linear transcript, which is derived from the same host gene. The ratios are presented against the abundance of the latter isoform. Note that most circRNAs are expressed at lower levels than the corresponding linear transcripts; only five circRNAs (red dots; identified by names) exceeded the abundance of their respective linear isoforms, but the absolute expression levels of all five circRNAs are relatively low.
Figure 3.
Analysis of newly transcribed RNAs reveals that circRNAs are more stable and static than the linear isoforms derived from the same host genes. (A) MCF10A cells were treated with EGF as in Figure 1B and RNA was simultaneously metabolically labeled using 4-thiouridine (4sU), for the indicated time intervals. RNA was extracted (Total-RNA) with Trizol, biotinylated and purified on streptavidin magnetic beads (denoted 4sU-RNA). Flow-through RNA was also collected (denoted FT-RNA). Thereafter, RNA was reverse transcribed and quantified using high-throughput real time PCR (Fluidigm). The boxplot diagram shows enrichment of newly transcribed RNA (4sU labeled) in linear isoforms (blue dots) relative to the respective circRNA isoforms (red dots; P < 1e-20, _t_-test, two-tailed distribution, unequal variance, Bonferroni corrected for multiple comparisons, N = 61). Shown are the log2 fold enrichments of 4sU-labeled RNA versus total RNA (Y axis). (B) MCF10A cells were metabolically labeled using 4sU, for 1 or 2 h. Thereafter, RNA was extracted, biotinylated and purified on streptavidin magnetic beads. Flow-through RNA was also collected. Next, RNA was reverse transcribed and quantified using high-throughput real time PCR (Fluidigm). Presented are the half-lives of 60 circRNAs and their corresponding linear counterparts. Half-life values were calculated from two samples, which were labeled with 4sU for 1 or 2 h and then averaged. All data were corrected for any bias introduced due to low uridine (short length) of RNA species (see ‘Materials and Methods’ section). The circRNAs (red dots) and their linear counterparts (blue dots) were sorted according to the difference between their half-lives from high to low. Error bars represent standard errors. The calculations of half life were performed using the HALO software (39) and the ratios between newly transcribed (RNA-4sU) and pre-existing RNAs (RNA-FT). (C) PCR and four pairs of primers were used to follow the response of newly transcribed RNA (solid lines) and total RNA (dashed lines) of both linear and circular isoforms of the indicated EGF-regulated genes. Shown are calculated fold changes of expression levels displayed by EGF-treated cells relative to basal (unstimulated) expression levels, measured in cells labeled with 4sU for the indicated intervals (Y axis). Shown are means ± S.E. of duplicates.
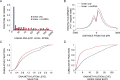
Figure 4.
Genes giving rise to circRNAs frequently express the respective mRNAs, and they are transcriptionally more active than genes transcribed into linear RNA only. (A) Basal (unstimulated) expression levels of mRNAs transcribed from genes that also give rise to circRNAs (red bars; N = 1051) are compared to mRNAs transcribed from genes that give rise only to linear RNAs (no detectable circRNAs; blue bars; N = 21 872). MCF10A RNA-seq data were obtained from a publicly available RNA-sequencing dataset (42). RPKM—reads per kilo basepairs per million reads. P = 6.13e-212, Mann–Whitney-U test. Note that only five circRNAs (leftmost quantiles) displayed no linear counterparts. (B) Basal H3K27 acetylation levels at promoter regions (transcription start site (TSS) ± 3000 bp) were determined in MCF10A cells. The acetylation levels displayed by genes transcribed into both circRNAs and linear RNAs (N = 781; red line) are compared to genes transcribed into linear RNAs with no indication of the respective circRNAs (N = 4992; blue line). The tag counts were normalized to 10 million sequencing tags for each sample. Shown are curves of tag densities of 25 nt bins. Tag density units are presented per basepair (bp) and per gene. Note that H3K27 acetylation levels are higher for genes producing circRNA compared to the control gene set (P = 1.26e-26, Mann–Whitney-U test). (C) Basal DNA methylation levels at the promoter region (TSS ± 1000 bp) of genes transcribed into both circRNAs and linear RNAs (N = 781) are compared to methylation levels displayed by genes that give rise to linear RNA only (N = 4992). DNA methylation data of MCF10A cells was derived from the ENCODE dataset. DNA methylation levels in the promoter region (−1000 to +1000 bp relative to the TSS) of each circRNA-producing host gene and control gene (no evidence for circRNA production) were tallied and a cumulative fraction was calculated and plotted for both groups. Note that DNA methylation levels are lower in genes producing also circRNAs compared to a group of genes giving rise to linear RNAs only (P = 3.15e-12, Mann–Whitney-U test). (D) Basal DNA methylation levels in the region between the TSS and TES (gene body) of genes transcribed into both circRNA and linear RNA (N = 399) are compared to levels of methylation displayed by genes that give rise to linear RNA with no detectable circRNAs (N = 2220). The methylation levels were tallied and normalized to gene length for each circRNA-producing host gene and for control genes (no evidence for circRNA production). A cumulative fraction was calculated and plotted for both groups. Note that DNA methylation levels in the body of genes producing circRNAs are lower compared to a control group of genes (P = 9.46e-57, Mann–Whitney-U test).
Figure 5.
Circular RNAs expressed by human mammary cells show no enrichment for binding sites of the microRNAs found in the same cells. (A) CircRNAs expressed in MCF10A cells were analyzed for the presence of binding sites (7 or 8 nt long) for miRNAs expressed by the same cells (33% uppermost percentile). The number of sites was tallied for each circRNA–miRNA pair, and the distribution of values is plotted. The solid and dotted curves indicate the averaged and 95% upper percentile, respectively, of results when repeating the analysis 1000 times using different permutations of site sequences (see ‘Materials and Methods’ section). (B) Shown are the numbers of hexamers within circRNAs of MCF10A cells that potentially pair to miRNAs expressed in the same cells (orange line), or to scrambled miRNA regions (black line). Error bars represent the distribution of values in 1000 control scrambled miRNA regions. Scrambled miRNA regions preserved mono-nucleotide and GC dinucleotide composition of the original 6 nt window. For each miRNA highly expressed in MCF10A cells (33% upper percentile), starting from the 5′ end, we checked 6-nt-long windows for complementarity to hexamers within the collection of circRNAs expressed in MCF10A cells (N = 1326). The total number of matches was normalized to the number of unique 6 nt windows analyzed. Highlighted is the central region of miRNAs found in a previous study to have statistically significant pairing to coding sequences (55).
Similar articles
- Amphiregulin is coordinately expressed with heparin-binding epidermal growth factor-like growth factor in the interstitial smooth muscle of the human prostate.
Adam RM, Borer JG, Williams J, Eastham JA, Loughlin KR, Freeman MR. Adam RM, et al. Endocrinology. 1999 Dec;140(12):5866-75. doi: 10.1210/endo.140.12.7221. Endocrinology. 1999. PMID: 10579352 - Blockage of EGF receptor signal transduction causes reversible arrest of normal and immortal human mammary epithelial cells with synchronous reentry into the cell cycle.
Stampfer MR, Pan CH, Hosoda J, Bartholomew J, Mendelsohn J, Yaswen P. Stampfer MR, et al. Exp Cell Res. 1993 Sep;208(1):175-88. doi: 10.1006/excr.1993.1236. Exp Cell Res. 1993. PMID: 7689475 - Altered gene expression of c-myc, epidermal growth factor receptor, transforming growth factor-alpha, and c-erb-B2 in an immortalized human breast epithelial cell line, HMT-3522, is associated with decreased growth factor requirements.
Madsen MW, Lykkesfeldt AE, Laursen I, Nielsen KV, Briand P. Madsen MW, et al. Cancer Res. 1992 Mar 1;52(5):1210-7. Cancer Res. 1992. PMID: 1737382 - The complexity of the translation ability of circRNAs.
Granados-Riveron JT, Aquino-Jarquin G. Granados-Riveron JT, et al. Biochim Biophys Acta. 2016 Oct;1859(10):1245-51. doi: 10.1016/j.bbagrm.2016.07.009. Epub 2016 Jul 19. Biochim Biophys Acta. 2016. PMID: 27449861 Review. - Circular RNA: A new star of noncoding RNAs.
Qu S, Yang X, Li X, Wang J, Gao Y, Shang R, Sun W, Dou K, Li H. Qu S, et al. Cancer Lett. 2015 Sep 1;365(2):141-8. doi: 10.1016/j.canlet.2015.06.003. Epub 2015 Jun 5. Cancer Lett. 2015. PMID: 26052092 Review.
Cited by
- CircPOLA2 sensitizes non-small cell lung cancer cells to ferroptosis and suppresses tumorigenesis via the Merlin-YAP signaling pathway.
Xu K, Wei G, Qi W, Ye C, Liu Y, Wang S, Yang F, Tang J. Xu K, et al. iScience. 2024 Aug 27;27(9):110832. doi: 10.1016/j.isci.2024.110832. eCollection 2024 Sep 20. iScience. 2024. PMID: 39310771 Free PMC article. - The Emerging Roles and Clinical Potential of circSMARCA5 in Cancer.
Xue C, Wei J, Li M, Chen S, Zheng L, Zhan Y, Duan Y, Deng H, Xiong W, Li G, Li H, Zhou M. Xue C, et al. Cells. 2022 Sep 30;11(19):3074. doi: 10.3390/cells11193074. Cells. 2022. PMID: 36231036 Free PMC article. Review. - Circular RNA vaccines with long-term lymph node-targeting delivery stability after lyophilization induce potent and persistent immune responses.
Wan J, Wang Z, Wang L, Wu L, Zhang C, Zhou M, Fu ZF, Zhao L. Wan J, et al. mBio. 2024 Jan 16;15(1):e0177523. doi: 10.1128/mbio.01775-23. Epub 2023 Dec 11. mBio. 2024. PMID: 38078742 Free PMC article. - CircRNA as an Achilles heel of cancer: characterization, biomarker and therapeutic modalities.
Zhang J, Luo Z, Zheng Y, Duan M, Qiu Z, Huang C. Zhang J, et al. J Transl Med. 2024 Aug 10;22(1):752. doi: 10.1186/s12967-024-05562-4. J Transl Med. 2024. PMID: 39127679 Free PMC article. Review. - RNA biomarkers for alcohol use disorder.
Ferguson LB, Mayfield RD, Messing RO. Ferguson LB, et al. Front Mol Neurosci. 2022 Nov 4;15:1032362. doi: 10.3389/fnmol.2022.1032362. eCollection 2022. Front Mol Neurosci. 2022. PMID: 36407766 Free PMC article. Review.
References
- Lo H.W., Hung M.C. Nuclear EGFR signalling network in cancers: linking EGFR pathway to cell cycle progression, nitric oxide pathway and patient survival. Br. J. Cancer. 2007;96(Suppl):R16–R20. - PubMed
- Ronnstrand L., Heldin C.H. Mechanisms of platelet-derived growth factor-induced chemotaxis. Int. J. Cancer. 2001;91:757–762. - PubMed
- Avraham R., Yarden Y. Feedback regulation of EGFR signalling: decision making by early and delayed loops. Nat. Rev. Mol. Cell Biol. 2011;12:104–117. - PubMed
- Avraham R., Sas-Chen A., Manor O., Steinfeld I., Shalgi R., Tarcic G., Bossel N., Zeisel A., Amit I., Zwang Y., et al. EGF decreases the abundance of microRNAs that restrain oncogenic transcription factors. Sci. Signal. 2010;3:ra43. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases