The Plastomes of Two Species in the Endoparasite Genus Pilostyles (Apodanthaceae) Each Retain Just Five or Six Possibly Functional Genes - PubMed (original) (raw)
The Plastomes of Two Species in the Endoparasite Genus Pilostyles (Apodanthaceae) Each Retain Just Five or Six Possibly Functional Genes
Sidonie Bellot et al. Genome Biol Evol. 2015.
Abstract
The 23 species of mycoheterotrophic or exoparasitic land plants (from 15 genera and 6 families) studied so far all retain a minimal set of 17 of the normally 116 plastome genes. Only Rafflesia lagascae, an endoparasite concealed in its host except when flowering, has been reported as perhaps lacking a plastome, although it still possesses plastid-like compartments. We analyzed two other endoparasites, the African Apodanthaceae Pilostyles aethiopica and the Australian Pilostyles hamiltonii, both living inside Fabaceae. Illumina and 454 data and Sanger resequencing yielded circularized plastomes of 11,348 and 15,167 bp length, with both species containing five possibly functional genes (accD, rps3, rps4, rrn16, rrn23) and two/three pseudogenes (rpoC2 in P. aethiopica and rpl2 and rps12 in both species; rps12 may be functional in P. hamiltonii). Previously known smallest land plant plastomes contain 27-29 genes, making these Apodanthaceae plastomes the most reduced in size and gene content. A similar extent of divergence might have caused the plastome of Rafflesia to escape detection. The higher plastome degeneration in both these families of endoparasites, Rafflesiaceae and Apodanthaceae, of similar high age, compared with exoparasites points to a difference of plastome function between those two modes of parasitic life.
Keywords: chloroplast genome; endoparasite; gene loss; minimal plastome; photosynthesis.
© The Author 2015. Published by Oxford University Press on behalf of the Society for Molecular Biology and Evolution.
Figures
Fig. 1.—
Flowers of Pilostyles aethiopica emerging from the host Julbernardia globiflora (Fabaceae) in Harare, Zimbabwe. Scale bar is 5 mm. Photo S. Bellot.
Fig. 2.—
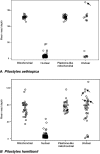
Read depth of Pilostyles contigs with plastome-like regions. Unclear: Contigs with plastome-like regions whose flanking regions were too short to infer their genomic location. Arrows indicate contigs that are part of plastomes (Results). The means for P. aethiopica are based on 34 contigs located in the chondriome, 670 located in the nuclear genome, 38 inferred to be in the chondriome from their flanks, and 19 of unclear location. The means for P. hamiltonii are based on 32 contigs located in the chondriome, 369 located in the nuclear genome, 31 inferred to be in the chondriome from their flanks, and 37 of unclear location.
Fig. 3.—
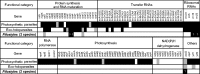
Map of the plastid genomes of P. aethiopica and P. hamiltonii. The skyline graphs represent the GC% with the minimum, mean, and maximum values indicated on the right. The blue and red bands indicate identity greater than 70% for bitscores greater than 100, red bands show a match in the same orientation whereas blue bands symbolize reversed-complement matches. The three bars above the gene labels refer to the reading frames; stop codons are represented by vertical black bars, and start codons (methionine) by purple vertical bars. Ψ means the gene is pseudogenized. Visualization obtained with the Artemis Comparison Tool (Carver et al. 2005).
Fig. 4.—
The plastome genes retained in parasitic and mycoheterotrophic land plants, based on Li et al. (2013), Barrett et al. (2014), Schelkunov et al. (2015), and Lam et al. (2015). White/grey fields indicate that a gene is lost/pseudogenized in at least one lineage of a given type of parasitism. Black fields indicate that a gene is functional in all lineages of the respective parasitism category. For Pilostyles, we did not take pseudogenized tRNAs into account, and the functioning of the genes remains to be confirmed (question marks).
Similar articles
- Plastome reduction and gene content in New World Pilostyles (Apodanthaceae) unveils high similarities to African and Australian congeners.
Arias-Agudelo LM, González F, Isaza JP, Alzate JF, Pabón-Mora N. Arias-Agudelo LM, et al. Mol Phylogenet Evol. 2019 Jun;135:193-202. doi: 10.1016/j.ympev.2019.03.014. Epub 2019 Mar 23. Mol Phylogenet Evol. 2019. PMID: 30914393 - Drastic reduction of plastome size in the mycoheterotrophic Thismia tentaculata relative to that of its autotrophic relative Tacca chantrieri.
Lim GS, Barrett CF, Pang CC, Davis JI. Lim GS, et al. Am J Bot. 2016 Jun;103(6):1129-37. doi: 10.3732/ajb.1600042. Epub 2016 Jun 22. Am J Bot. 2016. PMID: 27335389 - Investigating the path of plastid genome degradation in an early-transitional clade of heterotrophic orchids, and implications for heterotrophic angiosperms.
Barrett CF, Freudenstein JV, Li J, Mayfield-Jones DR, Perez L, Pires JC, Santos C. Barrett CF, et al. Mol Biol Evol. 2014 Dec;31(12):3095-112. doi: 10.1093/molbev/msu252. Epub 2014 Aug 28. Mol Biol Evol. 2014. PMID: 25172958 - Plastomes on the edge: the evolutionary breakdown of mycoheterotroph plastid genomes.
Graham SW, Lam VK, Merckx VS. Graham SW, et al. New Phytol. 2017 Apr;214(1):48-55. doi: 10.1111/nph.14398. Epub 2017 Jan 9. New Phytol. 2017. PMID: 28067952 Review. - Progress, challenge and prospect of plant plastome annotation.
Qu XJ, Zou D, Zhang RY, Stull GW, Yi TS. Qu XJ, et al. Front Plant Sci. 2023 May 30;14:1166140. doi: 10.3389/fpls.2023.1166140. eCollection 2023. Front Plant Sci. 2023. PMID: 37324662 Free PMC article. Review.
Cited by
- Comparative analysis of plastid genomes of non-photosynthetic Ericaceae and their photosynthetic relatives.
Logacheva MD, Schelkunov MI, Shtratnikova VY, Matveeva MV, Penin AA. Logacheva MD, et al. Sci Rep. 2016 Jul 25;6:30042. doi: 10.1038/srep30042. Sci Rep. 2016. PMID: 27452401 Free PMC article. - Comparative Analyses of 3,654 Plastid Genomes Unravel Insights Into Evolutionary Dynamics and Phylogenetic Discordance of Green Plants.
Yang T, Sahu SK, Yang L, Liu Y, Mu W, Liu X, Strube ML, Liu H, Zhong B. Yang T, et al. Front Plant Sci. 2022 Apr 11;13:808156. doi: 10.3389/fpls.2022.808156. eCollection 2022. Front Plant Sci. 2022. PMID: 35498716 Free PMC article. - Unprecedented Parallel Photosynthetic Losses in a Heterotrophic Orchid Genus.
Barrett CF, Sinn BT, Kennedy AH. Barrett CF, et al. Mol Biol Evol. 2019 Sep 1;36(9):1884-1901. doi: 10.1093/molbev/msz111. Mol Biol Evol. 2019. PMID: 31058965 Free PMC article. - The First Plastid Genome of the Holoparasitic Genus Prosopanche (Hydnoraceae).
Jost M, Naumann J, Rocamundi N, Cocucci AA, Wanke S. Jost M, et al. Plants (Basel). 2020 Mar 1;9(3):306. doi: 10.3390/plants9030306. Plants (Basel). 2020. PMID: 32121567 Free PMC article.
References
- Barbrook AC, Howe CJ, Purton S. 2006. Why are plastid genomes retained in non-photosynthetic organisms? Trends Plant Sci.. 11:101–108. - PubMed
- Barow M. 2006. Endopolyploidy in seed plants. Bioessays 28:271–281. - PubMed
- Barrett CF, Davis JI. 2012. The plastid genome of the mycoheterotrophic Corallorhiza striata (Orchidaceae) is in the relatively early stages of degradation. Am J Bot. 99:1513–1523. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous