Readers of poly(ADP-ribose): designed to be fit for purpose - PubMed (original) (raw)
Review
. 2016 Feb 18;44(3):993-1006.
doi: 10.1093/nar/gkv1383. Epub 2015 Dec 15.
Affiliations
- PMID: 26673700
- PMCID: PMC4756826
- DOI: 10.1093/nar/gkv1383
Review
Readers of poly(ADP-ribose): designed to be fit for purpose
Federico Teloni et al. Nucleic Acids Res. 2016.
Abstract
Post-translational modifications (PTMs) regulate many aspects of protein function and are indispensable for the spatio-temporal regulation of cellular processes. The proteome-wide identification of PTM targets has made significant progress in recent years, as has the characterization of their writers, readers, modifiers and erasers. One of the most elusive PTMs is poly(ADP-ribosyl)ation (PARylation), a nucleic acid-like PTM involved in chromatin dynamics, genome stability maintenance, transcription, cell metabolism and development. In this article, we provide an overview on our current understanding of the writers of this modification and their targets, as well as the enzymes that degrade and thereby modify and erase poly(ADP-ribose) (PAR). Since many cellular functions of PARylation are exerted through dynamic interactions of PAR-binding proteins with PAR, we discuss the readers of this modification and provide a synthesis of recent findings, which suggest that multiple structurally highly diverse reader modules, ranging from completely folded PAR-binding domains to intrinsically disordered sequence stretches, evolved as PAR effectors to carry out specific cellular functions.
© The Author(s) 2015. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures
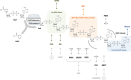
Figure 1.
Readers of poly(ADP-ribose). PAR polymerases use NAD+ to generate highly anionic linear and branched (not shown) PAR chains of different size and branching complexity. Besides the classical, well-characterized PAR reader modules WWE, PBZ, PBM, and macrodomains (top) also newly emerging PAR reader modules such as FHA, OB-fold, PIN domain, RRM, SR and KR repeats, RGG repeats and BRCT (bottom) appear as PAR readers and effectors. Multi-branched arrows indicate that the exact binding sites have not been defined.
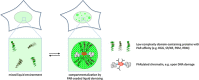
Figure 2.
PAR-seeded liquid demixing. PAR chains assemble hundreds of proteins, including many intrinsically disordered, low complexity domain-containing proteins, at sites of PAR formation, which collectively re-shape the local environment. This can lead to dynamic compartmentalization by liquid demixing, indicating that PAR can function as a general organizer of the soluble intracellular space.
Similar articles
- The recognition and removal of cellular poly(ADP-ribose) signals.
Barkauskaite E, Jankevicius G, Ladurner AG, Ahel I, Timinszky G. Barkauskaite E, et al. FEBS J. 2013 Aug;280(15):3491-507. doi: 10.1111/febs.12358. Epub 2013 Jun 18. FEBS J. 2013. PMID: 23711178 Review. - 50Years of poly(ADP-ribosyl)ation.
Virág L. Virág L. Mol Aspects Med. 2013 Dec;34(6):1043-5. doi: 10.1016/j.mam.2013.05.002. Epub 2013 May 28. Mol Aspects Med. 2013. PMID: 23727362 Review. - Poly(ADP-ribose): PARadigms and PARadoxes.
Bürkle A, Virág L. Bürkle A, et al. Mol Aspects Med. 2013 Dec;34(6):1046-65. doi: 10.1016/j.mam.2012.12.010. Epub 2013 Jan 2. Mol Aspects Med. 2013. PMID: 23290998 Review. - Poly(ADP-ribose) signaling in cell death.
Virág L, Robaszkiewicz A, Rodriguez-Vargas JM, Oliver FJ. Virág L, et al. Mol Aspects Med. 2013 Dec;34(6):1153-67. doi: 10.1016/j.mam.2013.01.007. Epub 2013 Feb 15. Mol Aspects Med. 2013. PMID: 23416893 Review. - Poly(ADP-ribose)-binding zinc finger motifs in DNA repair/checkpoint proteins.
Ahel I, Ahel D, Matsusaka T, Clark AJ, Pines J, Boulton SJ, West SC. Ahel I, et al. Nature. 2008 Jan 3;451(7174):81-5. doi: 10.1038/nature06420. Nature. 2008. PMID: 18172500
Cited by
- Poly(ADP-ribose) promotes toxicity of C9ORF72 arginine-rich dipeptide repeat proteins.
Gao J, Mewborne QT, Girdhar A, Sheth U, Coyne AN, Punathil R, Kang BG, Dasovich M, Veire A, DeJesus Hernandez M, Liu S, Shi Z, Dafinca R, Fouquerel E, Talbot K, Kam TI, Zhang YJ, Dickson D, Petrucelli L, van Blitterswijk M, Guo L, Dawson TM, Dawson VL, Leung AKL, Lloyd TE, Gendron TF, Rothstein JD, Zhang K. Gao J, et al. Sci Transl Med. 2022 Sep 14;14(662):eabq3215. doi: 10.1126/scitranslmed.abq3215. Epub 2022 Sep 14. Sci Transl Med. 2022. PMID: 36103513 Free PMC article. - Quantitative Analysis of Nuclear Poly(ADP-Ribose) Dynamics in Response to Laser-Induced DNA Damage.
Koczor CA, Saville KM, Al-Rahahleh RQ, Andrews JF, Li J, Sobol RW. Koczor CA, et al. Methods Mol Biol. 2023;2609:43-59. doi: 10.1007/978-1-0716-2891-1_3. Methods Mol Biol. 2023. PMID: 36515828 Free PMC article. - The key players of parthanatos: opportunities for targeting multiple levels in the therapy of parthanatos-based pathogenesis.
Liu L, Li J, Ke Y, Zeng X, Gao J, Ba X, Wang R. Liu L, et al. Cell Mol Life Sci. 2022 Jan 9;79(1):60. doi: 10.1007/s00018-021-04109-w. Cell Mol Life Sci. 2022. PMID: 35000037 Free PMC article. Review. - Role of microRNAs in regulation of doxorubicin and paclitaxel responses in lung tumor cells.
Maharati A, Moghbeli M. Maharati A, et al. Cell Div. 2023 Jul 21;18(1):11. doi: 10.1186/s13008-023-00093-8. Cell Div. 2023. PMID: 37480054 Free PMC article. Review. - Dynamic ADP-Ribosylome, Phosphoproteome, and Interactome in LPS-Activated Macrophages.
Daniels CM, Kaplan PR, Bishof I, Bradfield C, Tucholski T, Nuccio AG, Manes NP, Katz S, Fraser IDC, Nita-Lazar A. Daniels CM, et al. J Proteome Res. 2020 Sep 4;19(9):3716-3731. doi: 10.1021/acs.jproteome.0c00261. Epub 2020 Jul 1. J Proteome Res. 2020. PMID: 32529831 Free PMC article.
References
- Schreiber V., Dantzer F., Ame J.C., de Murcia G. Poly(ADP-ribose): novel functions for an old molecule. Nat. Rev. Mol. Cell Biol. 2006;7:517–528. - PubMed
- Burkle A. Poly(ADP-ribose). The most elaborate metabolite of NAD+ FEBS J. 2005;272:4576–4589. - PubMed
- Hassa P.O., Hottiger M.O. The diverse biological roles of mammalian PARPS, a small but powerful family of poly-ADP-ribose polymerases. Front. Biosci. 2008;13:3046–3082. - PubMed
- Barkauskaite E., Jankevicius G., Ladurner A.G., Ahel I., Timinszky G. The recognition and removal of cellular poly(ADP-ribose) signals. FEBS J. 2013;280:3491–3507. - PubMed
- Chambon P., Weill J.D., Mandel P. Nicotinamide mononucleotide activation of new DNA-dependent polyadenylic acid synthesizing nuclear enzyme. Bioch. Biophys. Res. Commun. 1963;11:39–43. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous