The prenucleosome, a stable conformational isomer of the nucleosome - PubMed (original) (raw)
The prenucleosome, a stable conformational isomer of the nucleosome
Jia Fei et al. Genes Dev. 2015.
Abstract
Chromatin comprises nucleosomes as well as nonnucleosomal histone-DNA particles. Prenucleosomes are rapidly formed histone-DNA particles that can be converted into canonical nucleosomes by a motor protein such as ACF. Here we show that the prenucleosome is a stable conformational isomer of the nucleosome. It consists of a histone octamer associated with ∼ 80 base pair (bp) of DNA, which is located at a position that corresponds to the central 80 bp of a nucleosome core particle. Monomeric prenucleosomes with free flanking DNA do not spontaneously fold into nucleosomes but can be converted into canonical nucleosomes by an ATP-driven motor protein such as ACF or Chd1. In addition, histone H3K56, which is located at the DNA entry and exit points of a canonical nucleosome, is specifically acetylated by p300 in prenucleosomes relative to nucleosomes. Prenucleosomes assembled in vitro exhibit properties that are strikingly similar to those of nonnucleosomal histone-DNA particles in the upstream region of active promoters in vivo. These findings suggest that the prenucleosome, the only known stable conformational isomer of the nucleosome, is related to nonnucleosomal histone-DNA species in the cell.
Keywords: active chromatin; gene expression; histones; nucleosome; prenucleosome.
© 2015 Fei et al.; Published by Cold Spring Harbor Laboratory Press.
Figures
Figure 1.
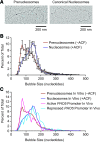
Psoralen cross-linking and electron microscopy analysis suggests that prenucleosomes associate with ∼70–80 bp of DNA. (A) Analysis of prenucleosomes and nucleosomes by psoralen cross-linking followed by denaturing electron microscopy. Representative images are shown. The histone-free DNA is cross-linked by psoralen, and the resulting bubbles represent the locations of prenucleosomes and nucleosomes. (B) ACF-mediated conversion of NAP1-assembled prenucleosomes to canonical nucleosomes increases the size of psoralen bubbles from ∼70–80 nt to ∼140–150 nt. Prenucleosomes (−ACF) and nucleosomes (+ACF) were subjected to psoralen cross-linking and denaturing electron microscopy. Bubble sizes were measured in ImageJ and converted from nanometers to nucleotides. A total of 4623 prenucleosome bubbles and 5013 nucleosome bubbles was measured in four independent experiments. The plot displays the distribution of bubble sizes as the average ± standard deviation (n = 4) of 10-nt bins. The individual data points are placed at the center of the 10-nt bins. (C) Comparison of the psoralen bubble sizes observed in vitro and in vivo. The data from prenucleosomes and nucleosomes in vitro (shown in B) and at the active versus repressed PHO5 promoters in vivo in Saccharomyces cerevisiae (Brown et al. 2013). The plot shows the distribution of bubble sizes as the average of 10-nt bins.
Figure 2.
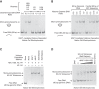
Rapid and efficient formation of mono-prenucleosomes with 80-bp DNA fragments. (A) The NAP1-mediated formation of mono-prenucleosomes with an 80-bp genomic DNA fragment occurs rapidly and requires all four core histones. Histone–NAP1 complexes were combined with an 80-bp DNA fragment (an 80-bp segment in the coding sequence of the Drosophila melanogaster ISWI gene; henceforth termed the “80-bp genomic DNA”). The samples were incubated at room temperature for 30 sec and then subjected to native (nondenaturing) 5% polyacrylamide gel electrophoresis. The DNA was visualized by staining with ethidium bromide. One octamer equivalent of all four core histones per DNA would be a 1:1 octamer:DNA ratio. Note that one octamer equivalent of H2A+H2B+H3+H4 has the same amount of H3 and H4 as one octamer equivalent of H3+H4. (B) Mono-prenucleosomes can be formed with the central 80 bp of the 601 nucleosome positioning sequence. Mono-prenucleosomes were formed and analyzed as in A, with all four core histones along with either the 80-bp genomic DNA or the central 80 bp of the 601 sequence. (C) Mono-prenucleosomes appear to be the thermodynamically most stable arrangement of the four core histones and 80 bp of DNA at 50 mM NaCl. Mono-prenucleosomes were formed with the dNLP histone chaperone as well as by salt dialysis of the four core histones with the 80-bp genomic DNA fragment. For comparison, H3–H4 monotetrasomes were also generated in parallel by salt dialysis with H3–H4. The histones were used at an octamer equivalent:DNA ratio of 1.0. (D) Mono-tetrasomes can be converted into prenucleosomes by the addition of H2A–H2B. Monotetrasomes were formed by salt dialysis with the 80 bp of genomic DNA as in C. Next, NAP1–H2A–H2B complexes were added as indicated. The samples were incubated for 30 sec at room temperature and then subjected to native 5% polyacrylamide gel electrophoresis. As a reference, a mono-prenucleosome formed by salt dialysis as in C was included (“Mono-prenuc”).
Figure 3.
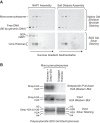
Mono-prenucleosomes contain all four core histones and are distinct from hexasomes. (A) Sucrose gradient sedimentation analysis reveals that mono-prenucleosomes contain all four core histones. Mono-prenucleosomes were prepared by either NAP1-mediated deposition (left panels) or salt dialysis (right panels) and then subjected to 10%–30% (w/v) (left to right) sucrose gradient sedimentation in a Beckman SW41 rotor (32,000 rpm for 18 h at 4°C). The arrows indicate the direction of sedimentation. (Top panels) The presence of mono-prenucleosomes was detected by native polyacrylamide gel electrophoresis and ethidium bromide staining of the DNA. (Bottom panels) The protein composition was analyzed by SDS–polyacrylamide gel electrophoresis and silver staining. The top two fractions and the bottom fraction did not contain histones (for example, see Supplemental Fig. S2A) and are not included. The sedimentation of the free core histones relative to prenucleosomal histones is shown in Supplemental Figure S2A. (B) Mono-prenucleosomes contain two copies of H2A and thus appear to contain a core histone octamer rather than hexamer. Mono-prenucleosomes were reconstituted with recombinant core histones onto the 80-bp genomic DNA by NAP1-mediated histone deposition. The H2A species were a combination of wild-type H2A and Strep-H2A at a 3:1 ratio of H2A:Strep-H2A. Prenucleosomes containing Strep-H2A were pulled down with streptavidin beads and then analyzed by Western blot with antibodies against histone H2A. An H2A Western blot and silver-stained SDS gel are also shown for the input samples. The Western blots were detected and quantitated by using 32P-labeled protein A.
Figure 4.
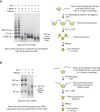
Prenucleosomes can be converted into canonical nucleosomes by ACF. (A) ACF-dependent assembly of poly-prenucleosomes to polynucleosomes. Mono-prenucleosomes (prepared by salt dialysis with 80-bp genomic DNA containing two 5-nt overhangs) were ligated in a sequential head-to-tail fashion with free DNA (90 bp with two 5-nt overhangs) to give poly-prenucleosomes, as indicated in the diagram. The resulting poly-prenucleosomes were assembled into polynucleosomes with ACF and ATP. The formation of canonical nucleosomes was verified by MNase digestion of the polynucleosomes into core particles, which contain ∼147 bp of DNA. (B) ACF-dependent assembly of mono-prenucleosomes to canonical nucleosomes. Mono-prenucleosomes (prepared by NAP1-mediated histone deposition with the central 80 bp of 601 DNA containing two 5-nt overhangs) were ligated to two free 80-bp DNA fragments (each containing a single 5-nt overhang) to give mono-prenucleosomes that are flanked by 85-bp DNA extensions, as illustrated in the diagram. The resulting mono-prenucleosomes were assembled into nucleosomes by ACF. The formation of canonical nucleosomes was assessed by MNase digestion into core particles that contain ∼147 bp of DNA. The 80-bp and 165-bp DNA fragments are incomplete ligation products.
Figure 5.
MNase digestion analysis reveals that prenucleosomes are associated with ∼80 bp of DNA. Mono-prenucleosomes were reconstituted by NAP1-mediated deposition onto the central 80 bp of the 601 DNA (with two 5-nt overhangs) and ligated to two free 80-bp DNA fragments (each containing a single 5-nt overhang) to give mono-prenucleosomes that are flanked by 85-bp DNA extensions, as indicated in the diagram. The samples were digested with MNase, and the 5′ ends of the resulting DNA fragments were mapped by primer extension analysis. The 5′ ends of the primers corresponded to the ends of the central 80 bp of the 601 sequences; thus, the majority of the MNase-digested fragments ranged in size from ∼78 to 85 bp. The control lanes show the ends of the unligated (single white dots) and ligated (double white dots) DNA fragments prior to MNase digestion.
Figure 6.
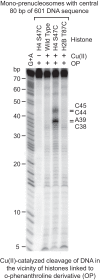
Mapping of the histone–DNA contacts in a mono-prenucleosome. Core histones containing the wild-type or the indicated mutant histones were modified with OP, which links an o-phenanthroline moiety onto the histones via alkylation of the thiol group on cysteine residues. The resulting derivatized histones were reconstituted by salt dialysis into mono-prenucleosomes with the central 80 bp of 601 DNA that is 32P-labeled at the 5′ end. The hydroxyl radical cleavage reactions were initiated by the addition of Cu(II), hydrogen peroxide, and mercaptopropionic acid. The cleavage products were purified and analyzed by electrophoresis on a 10% polyacrylamide–urea gel. The Maxam-Gilbert G+A ladder was used to identify the OP cleavage products, which are indicated at the right side of the autoradiogram. In a canonical nucleosome, H4S47C is located near the dyad.
Figure 7.
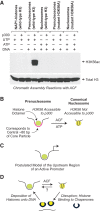
The prenucleosome, a conformational isomer of the nucleosome. (A) p300 specifically acetylates histone H3K56 in prenucleosomes relative to nucleosomes. Chromatin assembly reactions with ACF (Fyodorov and Kadonaga 2003; Torigoe et al. 2011) were performed with relaxed circular plasmid DNA in the presence of acetyl-CoA. ATP (or UTP as the −ATP control), DNA, and p300 were included as indicated. In addition, as a test for acetylation at H3K56, we performed parallel reactions with the mutant histone H3K56A, which cannot be acetylated at H3 residue 56. The resulting samples were then subjected to Western blot analysis with H3K56ac-specific antibodies (Millipore, catalog no. 07-677). As a reference, the blot was stripped and reprobed with anti-total H3 antibodies (Abcam, catalog no. AB1791). (B) Prenucleosomes comprise a core histone octamer and 80 bp of DNA at a location that is analogous to that of the central 80 bp of the core particle. H3K56 is accessible to p300 in a prenucleosome but not a nucleosome. Prenucleosomes can be converted into canonical nucleosomes by a motor protein such as ACF or Chd1. (C) Prenucleosomes or prenucleosome-related particles may be present in the upstream region of active promoters. (D) Model for the productive dynamic interconversion between prenucleosomes and nucleosomes. Prenucleosomes can be formed by the deposition of histones onto DNA and converted into nucleosomes by an ATP-driven motor protein such as ACF or Chd1. Nucleosomes can be disrupted by the action of enzymes such as polymerases as well as some ATP-driven chromatin remodeling factors. The resulting free histones are bound by the chaperones and then reassembled into prenucleosomes. It is not known whether a canonical nucleosome can be directly converted into a prenucleosome.
Similar articles
- Prenucleosomes and Active Chromatin.
Khuong MT, Fei J, Ishii H, Kadonaga JT. Khuong MT, et al. Cold Spring Harb Symp Quant Biol. 2015;80:65-72. doi: 10.1101/sqb.2015.80.027300. Epub 2016 Jan 14. Cold Spring Harb Symp Quant Biol. 2015. PMID: 26767995 Free PMC article. Review. - Identification of a rapidly formed nonnucleosomal histone-DNA intermediate that is converted into chromatin by ACF.
Torigoe SE, Urwin DL, Ishii H, Smith DE, Kadonaga JT. Torigoe SE, et al. Mol Cell. 2011 Aug 19;43(4):638-48. doi: 10.1016/j.molcel.2011.07.017. Mol Cell. 2011. PMID: 21855802 Free PMC article. - Distinct activities of CHD1 and ACF in ATP-dependent chromatin assembly.
Lusser A, Urwin DL, Kadonaga JT. Lusser A, et al. Nat Struct Mol Biol. 2005 Feb;12(2):160-6. doi: 10.1038/nsmb884. Epub 2005 Jan 9. Nat Struct Mol Biol. 2005. PMID: 15643425 - Chromatin modification by PSC occurs at one PSC per nucleosome and does not require the acidic patch of histone H2A.
Lo SM, McElroy KA, Francis NJ. Lo SM, et al. PLoS One. 2012;7(10):e47162. doi: 10.1371/journal.pone.0047162. Epub 2012 Oct 11. PLoS One. 2012. PMID: 23071745 Free PMC article. - DNA binding within the nucleosome core.
Luger K, Richmond TJ. Luger K, et al. Curr Opin Struct Biol. 1998 Feb;8(1):33-40. doi: 10.1016/s0959-440x(98)80007-9. Curr Opin Struct Biol. 1998. PMID: 9519294 Review.
Cited by
- Enhancement of homology-directed repair with chromatin donor templates in cells.
Cruz-Becerra G, Kadonaga JT. Cruz-Becerra G, et al. Elife. 2020 Apr 28;9:e55780. doi: 10.7554/eLife.55780. Elife. 2020. PMID: 32343230 Free PMC article. - NDF, a nucleosome-destabilizing factor that facilitates transcription through nucleosomes.
Fei J, Ishii H, Hoeksema MA, Meitinger F, Kassavetis GA, Glass CK, Ren B, Kadonaga JT. Fei J, et al. Genes Dev. 2018 May 1;32(9-10):682-694. doi: 10.1101/gad.313973.118. Epub 2018 May 14. Genes Dev. 2018. PMID: 29759984 Free PMC article. - Jim Kadonaga: Exploring transcription and chromatin.
Sedwick C. Sedwick C. J Cell Biol. 2016 Mar 14;212(6):608-9. doi: 10.1083/jcb.2126pi. J Cell Biol. 2016. PMID: 26975847 Free PMC article. - Parallel mapping with site-directed hydroxyl radicals and micrococcal nuclease reveals structural features of positioned nucleosomes in vivo.
Fuse T, Katsumata K, Morohoshi K, Mukai Y, Ichikawa Y, Kurumizaka H, Yanagida A, Urano T, Kato H, Shimizu M. Fuse T, et al. PLoS One. 2017 Oct 26;12(10):e0186974. doi: 10.1371/journal.pone.0186974. eCollection 2017. PLoS One. 2017. PMID: 29073207 Free PMC article. - H3.3 kinetics predicts chromatin compaction status of parental genomes in early embryos.
Guo SM, Liu XP, Zhou LQ. Guo SM, et al. Reprod Biol Endocrinol. 2021 Jun 11;19(1):87. doi: 10.1186/s12958-021-00776-3. Reprod Biol Endocrinol. 2021. PMID: 34116678 Free PMC article.
References
- Arimura Y, Tachiwana H, Oda T, Sato M, Kurumizaka H. 2012. Structural analysis of the hexasome, lacking one histone H2A/H2B dimer from the conventional nucleosome. Biochemistry 51: 3302–3309. - PubMed
- Brown CR, Eskin JA, Hamperl S, Griesenbeck J, Jurica MS, Boeger H. 2015. Chromatin structure analysis of single gene molecules by psoralen cross-linking and electron microscopy. Methods Mol Biol 1228: 93–121. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous