Evidence supporting a critical contribution of intrinsically disordered regions to the biochemical behavior of full-length human HP1γ - PubMed (original) (raw)
Evidence supporting a critical contribution of intrinsically disordered regions to the biochemical behavior of full-length human HP1γ
Gabriel Velez et al. J Mol Model. 2016 Jan.
Abstract
HP1γ, a non-histone chromatin protein, has elicited significant attention because of its role in gene silencing, elongation, splicing, DNA repair, cell growth, differentiation, and many other cancer-associated processes, including therapy resistance. These characteristics make it an ideal target for developing small drugs for both mechanistic experimentation and potential therapies. While high-resolution structures of the two globular regions of HP1γ, the chromo- and chromoshadow domains, have been solved, little is currently known about the conformational behavior of the full-length protein. Consequently, in the current study, we use threading, homology-based molecular modeling, molecular mechanics calculations, and molecular dynamics simulations to develop models that allow us to infer properties of full-length HP1γ at an atomic resolution level. HP1γ appears as an elongated molecule in which three Intrinsically Disordered Regions (IDRs, 1, 2, and 3) endow this protein with dynamic flexibility, intermolecular recognition properties, and the ability to integrate signals from various intracellular pathways. Our modeling also suggests that the dynamic flexibility imparted to HP1γ by the three IDRs is important for linking nucleosomes with PXVXL motif-containing proteins, in a chromatin environment. The importance of the IDRs in intermolecular recognition is illustrated by the building and study of both IDR2 HP1γ-importin-α and IDR1 and IDR2 HP1γ-DNA complexes. The ability of the three IDRs for integrating cell signals is demonstrated by combined linear motif analyses and molecular dynamics simulations showing that posttranslational modifications can generate a histone mimetic sequence within the IDR2 of HP1γ, which when bound by the chromodomain can lead to an autoinhibited state. Combined, these data underscore the importance of IDRs 1, 2, and 3 in defining the structural and dynamic properties of HP1γ, discoveries that have both mechanistic and potentially biomedical relevance.
Keywords: CBX3; Chromatin; Epigenetics; HP1; HP1γ; Molecular dynamics; Molecular modeling.
Figures
Fig. 1
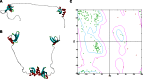
Structural bioinformatics reveal the HP1γ IDR regions to have a high propensity toward disorder. a Linear domain graph showing amino acid positions for IDR1 (Met1 – Val31), IDR2 (Ala82 – Arg115), and IDR3 (Arg171 – Gln183). b Hydrophobicity plot reveals the IDRs have a high polar to hydrophobic ratio of residues, a feature that characterizes intrinsically disordered proteins. c Disorder meta-prediction for full-length HP1γ reveals the IDRs to have a higher propensity toward disorder, while the chromo and chromoshadow domain are predicted to be ordered regions of the protein. Disorder probability values above the cut-off value of 0.5 are considered to be disordered. d Structure predictions of IDR1 e, IDR2, and IDR3 f predicted by threading algorithms show that threading cannot model these regions in any secondary or tertiary conformation
Fig. 2
Constructed models of the HP1γ monomer and homodimer. a Full-length model of HP1γ was generated by joining the globular domains (CD and CSD) with the IDRs using the Builder function in Discovery Studio Client 4.0. b Constructed model of the HP1γ homodimer. c Ramachandran plot reveals 97 % of the residues for this model to be in favored or allowed regions
Fig. 3
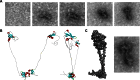
Electron microscopy validates the shape of the HP1γ homodimer. a Electron microscopy (EM) images of purified HP1γ. b Homology-based model of the HP1γ homodimer. c Superimposition of the homodimer structure shows that the predicted model fits nicely with the shape determined by EM imaging. Similar observations have been recently obtained for the yeast HP1 proteins, SWI6 [9]
Fig. 4
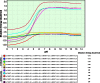
Molecular dynamics simulations of the HP1γ monomer. In order to gain further insight into the biochemical behavior of HP1γ, we subsequently utilized molecular dynamics simulations to perform a conformational search that might reflect the biophysical behavior of HP1 proteins. a An assemblage of conformers obtained during a short 100-ps MD simulation. Analysis of the trajectories obtained through this approach reveals that the highly flexible linker has the ability to shorten the length of this protein very rapidly. b A numerical representation of the flexibility and mobility of this protein during the simulation time was obtained by calculating the root-mean-square deviation (RMSD) and c root-mean-square fluctuation (RMSF). d Conformational sampling of the 2-ns GB model simulation highlights the same characteristic flexibility of IDR2 as the implicit solvent model. These results are congruent with the structural bioinformatics analyses, which suggest that the linker region has the highest propensity toward disorder. e, f RMSD and RMSF values for the generalized born (GB) simulation are also represented. g Radius of gyration calculation for the generalized born (GB) simulation
Fig. 5
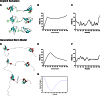
Identification of a nuclear localization signal in HP1γ. a Sequence comparison of HP1 NLS motifs with other validated NLS sequences was performed with MUSCLE [76]. This comparison validates the presence of these motifs in these proteins. b Since no mutational analyses exist that provide clues as to the cellular and molecular function of this region of the HP1 proteins, we applied the PsortII algorithm to determine that they primarily form a bipartite NLS that conforms to the consensus sequence (K/R)(K/R)_X_10–12(K/R)3/5, where (K/R)3/5 represents at least three of either lysine or arginine of five consecutive amino acids. The fact that the linkers of HP1 proteins are primarily composed of NLS motifs is in agreement with the results of our sequence-to-structural predictions, since all structural studies performed to date for this type of domains reveal their high degree of flexibility and tendency to disorder. c Immunoprecipitation of HP1γ followed by mass spectrometry demonstrated that this protein co-purifies in complex with the NLS receptor proteins, α-importins
Fig. 6
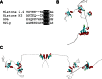
Modeling and simulation of the HP1γ-α-importin complex. a Model of IDR docked to the binding site of α-importin. The minor site specifically binds to the N-terminal basic cluster KR (represented in blue), and the larger, C-terminal basic cluster KRKK (represented in blue) binds to the major site. b Model of HP1γ bound to α-importin. The steric environments created by this interaction leaves room for both the chromodomain and chromoshadowdomain of HP1γ to extend outward and downward from the intermolecular interphase. c Molecular dynamics simulation of the complex. Unlike the isolated HP1γ simulation, binding to α-importin greatly restricts the disordered motion of IDR2
Fig. 7
Modeling of HP1γ–DNA complexes. a Prediction of DNA-binding residues by DP-Bind. Results of SVM, KLR, and PLR are represented along with the majority or consensus score of the three predictions, showing DNA-binding residues in the three IDR regions. b HP1γ–DNA complex generated by DP-Dock. Note how the interaction of IDR2 with B-DNA is consistent with the results of the DP-Bind prediction. c Model of IDR1 bound to a single nucleosome. d Model of the HP1γ dimer docked to two nucleosomes. e MD simulation of the HP1-nucleosome complex shows the PXVXL-domain docked peptide recruited by the HP1γ dimer. The most N-terminal domain of HP1γ contacts the DNA, which is in agreement with experimental data
Fig. 8
Characterization of IDR2 as the signal integration center through prediction of post-translational modifications. Prediction of post-translational modification sites on HP1γ was performed by compiling and statistically scoring linear motifs for phosphorylation (a), acetylation (b), methylation (c), ubiquitination (d), and sumoylation (e) as predicted by 20 different software programs. For each program, we considered sites for which the prediction score was above the cut-off that had been derived using a training set of modified sequences that have been experimentally validated. Subsequently, we developed a meta-prediction score by assigning a maximum score of 1 to sites that were predicted by all of the programs cited. Scores for other programs were numerically expressed relative to this maximum score of 1. This analysis revealed that predicted phosphorylation sites have high specificity potential near IDR2 of HP1γ. f Graphical representation of predicted post-translational modification sites. Results of this analysis revealed that post-translational modifications have the propensity to occur throughout the entire HP1γ sequence, but appear to be heavily localized to the IDR regions. g Graphical representation of experimentally validated post-translational modification sites listed on PhosphositePlus [57] and PHOSIDA [58]
Fig. 9
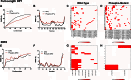
Effects of mutation on the intramolecular binding properties of HP1γ. Molecular dynamics simulations were used to determine the effect of phosphorylation on the stability and intramolecular binding of HP1γ. a, b Comparative MD simulations of the full-length wild-type HP1γ and phosphorylated HP1γ with the following amino acid positions mutated: 55, 60, 79, 89, 93, 95, 97, 99, 102, and 176. c, d The simulation results suggest that phosphorylation increases the time-dependent intramolecular binding of the phosphorylated mutant when compared to the wild type. e, f Comparative MD simulations of the full-length wild-type IDR3 and phosphorylated IDR3 with the following amino acid positions mutated: 169 and 172. g, h Similar to the full-length HP1γ, these simulation results suggest that phosphorylation increases the time-dependent intramolecular binding of the phosphorylated mutant when compared to the wild type. Together, these results support the use of linear motif analysis to predict post-translational modifications as these simulations suggest a relevant biophysical effect of phosphorylation on the behavior and intramolecular binding of HP1γ
Fig. 10
Combinatorial amino acid scanning mutagenesis reveals effect of phosphorylation on the binding affinity between HP1γ and α-importin. Residues in IDR2 were changed to either glutamic acid (phospho-mimicking) or alanine (non-phosphorylatable). Calculated mutation energies that were less than −0.5 kcal/mol were considered stabilizing while energies greater than 0.5 kcal/mol were considered destabilizing. Phospho-mimicking mutations displayed lower mutation energy profiles and thus a lower binding affinity for α-importin. Results of this analysis revealed that phosphorylation-mimicking mutations decrease affinity of HP1 for α-importin
Fig. 11
Modeling of the HP1γ auto-inhibited state. a Sequence alignment of the histone mimetic peptide of HP1γ (K82) with Histone 3 (K9), G9a (K185), and Histone 1.4 (K26). b Homology-based model of HP1γ in its autoinhibited state. c Homology-based model of the autoinhibited HP1γ homodimer. Together, these highlight the hypothesis that the ability of HP1γ to function as a histone mark reader is inhibited when both of its chromodomains are used to bind to additional monomers
Similar articles
- Discovery, expression, cellular localization, and molecular properties of a novel, alternative spliced HP1γ isoform, lacking the chromoshadow domain.
Mathison A, Milech De Assuncao T, Dsouza NR, Williams M, Zimmermann MT, Urrutia R, Lomberk G. Mathison A, et al. PLoS One. 2020 Feb 6;15(2):e0217452. doi: 10.1371/journal.pone.0217452. eCollection 2020. PLoS One. 2020. PMID: 32027651 Free PMC article. - Peptide recognition by heterochromatin protein 1 (HP1) chromoshadow domains revisited: Plasticity in the pseudosymmetric histone binding site of human HP1.
Liu Y, Qin S, Lei M, Tempel W, Zhang Y, Loppnau P, Li Y, Min J. Liu Y, et al. J Biol Chem. 2017 Apr 7;292(14):5655-5664. doi: 10.1074/jbc.M116.768374. Epub 2017 Feb 21. J Biol Chem. 2017. PMID: 28223359 Free PMC article. - Conformational diversity of human HP1α.
Ukmar-Godec T, Yu T, de Opakua AI, Pantoja CF, Munari F, Zweckstetter M. Ukmar-Godec T, et al. Protein Sci. 2024 Jul;33(7):e5079. doi: 10.1002/pro.5079. Protein Sci. 2024. PMID: 38895997 Free PMC article. - Structural features and interfacial properties of WH2, β-thymosin domains and other intrinsically disordered domains in the regulation of actin cytoskeleton dynamics.
Renault L, Deville C, van Heijenoort C. Renault L, et al. Cytoskeleton (Hoboken). 2013 Nov;70(11):686-705. doi: 10.1002/cm.21140. Epub 2013 Oct 22. Cytoskeleton (Hoboken). 2013. PMID: 24027208 Review. - Biochemical and structural properties of heterochromatin protein 1: understanding its role in chromatin assembly.
Nishibuchi G, Nakayama J. Nishibuchi G, et al. J Biochem. 2014 Jul;156(1):11-20. doi: 10.1093/jb/mvu032. Epub 2014 May 13. J Biochem. 2014. PMID: 24825911 Review.
Cited by
- Modeling post-translational modifications and cancer-associated mutations that impact the heterochromatin protein 1α-importin α heterodimers.
Zimmermann MT, Williams MM, Klee EW, Lomberk GA, Urrutia R. Zimmermann MT, et al. Proteins. 2019 Nov;87(11):904-916. doi: 10.1002/prot.25752. Epub 2019 Jun 14. Proteins. 2019. PMID: 31152607 Free PMC article. - CBX3/HP1γ promotes tumor proliferation and predicts poor survival in hepatocellular carcinoma.
Zhong X, Kan A, Zhang W, Zhou J, Zhang H, Chen J, Tang S. Zhong X, et al. Aging (Albany NY). 2019 Aug 2;11(15):5483-5497. doi: 10.18632/aging.102132. Epub 2019 Aug 2. Aging (Albany NY). 2019. PMID: 31375643 Free PMC article. - Elucidation of _MRAS_-mediated Noonan syndrome with cardiac hypertrophy.
Higgins EM, Bos JM, Mason-Suares H, Tester DJ, Ackerman JP, MacRae CA, Sol-Church K, Gripp KW, Urrutia R, Ackerman MJ. Higgins EM, et al. JCI Insight. 2017 Mar 9;2(5):e91225. doi: 10.1172/jci.insight.91225. JCI Insight. 2017. PMID: 28289718 Free PMC article. - Liquid droplet formation by HP1α suggests a role for phase separation in heterochromatin.
Larson AG, Elnatan D, Keenen MM, Trnka MJ, Johnston JB, Burlingame AL, Agard DA, Redding S, Narlikar GJ. Larson AG, et al. Nature. 2017 Jul 13;547(7662):236-240. doi: 10.1038/nature22822. Epub 2017 Jun 21. Nature. 2017. PMID: 28636604 Free PMC article. - Evolutional heterochromatin condensation delineates chromocenter formation and retrotransposon silencing in plants.
Zhang W, Cheng L, Li K, Xie L, Ji J, Lei X, Jiang A, Chen C, Li H, Li P, Sun Q. Zhang W, et al. Nat Plants. 2024 Aug;10(8):1215-1230. doi: 10.1038/s41477-024-01746-4. Epub 2024 Jul 16. Nat Plants. 2024. PMID: 39014153
References
- Eissenberg JC, James T, Foster-Hartnett DM, Hartnett T, Ngan V, Elgin SC. Mutation in a heterochromatin-specific chromosomal protein is associated with suppression of position-effect variegation in Drosophila melanogaster. Proc Natl Acad Sci U S A. 1990;87:9923–9927. doi: 10.1073/pnas.87.24.9923. - DOI - PMC - PubMed
- Velez G, Urrutia R, Lomberk G. Critical role of the HP1-histone methyl transferase pathways in cancer epigenetics. Med Epigenet. 2013;1(1):100–105. doi: 10.1159/000355978. - DOI
Publication types
MeSH terms
Substances
Grants and funding
- R01 CA178627/CA/NCI NIH HHS/United States
- R01 AI-089714/AI/NIAID NIH HHS/United States
- R01 DK052913/DK/NIDDK NIH HHS/United States
- R01 DK52913/DK/NIDDK NIH HHS/United States
- R01 AI089714/AI/NIAID NIH HHS/United States
- P50 CA102701/CA/NCI NIH HHS/United States
- P30DK084567/DK/NIDDK NIH HHS/United States
- T32 GM007337/GM/NIGMS NIH HHS/United States
- P30 DK084567/DK/NIDDK NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous