The nuclear basket mediates perinuclear mRNA scanning in budding yeast - PubMed (original) (raw)
The nuclear basket mediates perinuclear mRNA scanning in budding yeast
Mark-Albert Saroufim et al. J Cell Biol. 2015.
Abstract
After synthesis and transit through the nucleus, messenger RNAs (mRNAs) are exported to the cytoplasm through the nuclear pore complex (NPC). At the NPC, messenger ribonucleoproteins (mRNPs) first encounter the nuclear basket where mRNP rearrangements are thought to allow access to the transport channel. Here, we use single mRNA resolution live cell microscopy and subdiffraction particle tracking to follow individual mRNAs on their path toward the cytoplasm. We show that when reaching the nuclear periphery, RNAs are not immediately exported but scan along the nuclear periphery, likely to find a nuclear pore allowing export. Deletion or mutation of the nuclear basket proteins MLP1/2 or the mRNA binding protein Nab2 changes the scanning behavior of mRNPs at the nuclear periphery, shortens residency time at nuclear pores, and results in frequent release of mRNAs back into the nucleoplasm. These observations suggest a role for the nuclear basket in providing an interaction platform that keeps RNAs at the periphery, possibly to allow mRNP rearrangements before export.
© 2015 Saroufim et al.
Figures
Figure 1.
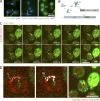
mRNAs scan the nuclear periphery before export to the cytoplasm. (A) Localization of CLB2, MDN1, and polyA mRNA by FISH suggests a rate-limiting step at the nuclear periphery. Blue arrows show sites of transcription. Yellow arrows show single mRNAs. DNA is stained with DAPI (blue). (B) Cartoon illustrating the mRNA labeling strategy using PP7 stem loops. (C) Live cell imaging of CLB2 mRNA. Individual frames from video acquired in 37-ms intervals. MAX shows the maximum intensity projection of all frames. mRNA is shown in green and NPC in red. The yellow arrows show single tracked mRNA in each frame. The purple arrow outlines the same RNA path in the MAX projected image. (D) Track of nuclear CLB2 mRNA from C and
Video 1
. The RNA position in each frame was determined using 2D Gaussian fitting and was superimposed onto a single frame of Video 1 (middle). Left panel shows connected positions to visualize the path from the nucleus to the cytoplasm.
Figure 2.
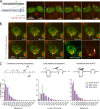
Jump distances of scanning RNAs suggest movement between pores. (A) Kinetics of 24PP7-GLT1 mRNA expression upon induction by galactose. Cartoon outlining the GAL1pro-24PP7-GLT1 reporter (left). Single images of 24PP7-GLT1 mRNA (green) and NPC (red) for indicated time points after the addition of galactose (right). Blue arrows show sites of transcription where multiple nascent mRNAs are associated with the GAL1pro-24PP7-GLT1 locus. Yellow arrows show single mRNAs. (B) Live cell tracking of scanning 24PP7-GLT1 mRNAs. Individual frames from a video acquired in 37-ms intervals. Lower right panel shows all nuclear positions of a single RNA superimposed onto a single frame. Blue arrows show sites of transcription, and yellow and purple arrows show the tracked mRNA in individual frames and MAX projected image, respectively. (C) Characterization of perinuclear mRNA scanning for CLB2-12xPP7, MDN1-12xPP7, and GAL1pro-24PP7-GLT1 mRNAs. Timescales of continuous mRNP scanning (left), jump distance at the periphery (middle), and timescales of restricted movements (right) are shown. Jump distances for the GAL1pro-24PP7-GLT1 transcription sites are shown in green. 156 (GLT1), 171 (MDN1), and 104 (CLB2) tracks were analyzed. See text for details.
Figure 3.
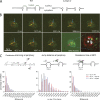
The nuclear basket is required for perinuclear scanning. (A) Cartoon illustrating the phenotype of mlp1/2 deletion leading to basketless pores. (B) Live cell RNA imaging of GAL1pro-24PP7-GLT1 mRNA shows reduced residence time at the nuclear periphery. Individual frames from a video acquired in 37-ms intervals. Lower right panel shows all nuclear positions of a single RNA superimposed onto a single frame. Blue arrows show sites of transcription, and yellow and purple arrows show the tracked mRNA in individual frames and MAX projected image, respectively. (C) Characterization of perinuclear mRNA scanning for GAL1pro-24PP7-GLT1 mRNAs in WT Δmlp1/2 and Δnup60. Timescales of continuous mRNP scanning (left), jump distance at the periphery (middle), and timescales of restricted movements (right) are shown. 156 (WT), 105 (ΔMLP1/2), and 76 (ΔNUP60) tracks were analyzed. See text for details. P < 0.05, comparing WT versus mutants using a randomized ANOVA followed by posthoc tests.
Figure 4.
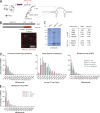
mRNP–NPC interactions mediate perinuclear scanning. (A) Cartoon describing the relationship between the nuclear polyA RNA binding protein Nab2 and the C-terminal domain of Mlp1p. (B) Localization of the Mlp1ΔC-2xmCherry fusion protein to the nuclear periphery. See text for details. (C) Deletion of the C terminus of Mlp1 does not affect basket integrity. Coomassie-stained gel separating protein complexes isolated by single-step affinity purification using Mlp1-ProtA, Mlp1ΔC-ProtA, or ProtA as baits. White line indicates that intervening lanes have been spliced out. Table with normalized peptide counts of copurified proteins as determined by mass spectrometry. Only selected NPC components are shown; for full list, see
Table S2
. (D) Quantification of GAL1pro-24PP7-GLT1 mRNP scanning behavior in mlp1ΔC and nab2F73D. 156 (WT), 75 (Mlp1-ΔC), and 85 (Nab2 F73D) tracks were analyzed. (E) Frequency of static frames at the periphery for GAL1pro-24PP7-GLT1 mRNPs in Δtom1 strain. P < 0.05, comparing WT versus mutants using a randomized ANOVA followed by posthoc tests, except WT versus Mlp1ΔC for scanning.
Similar articles
- In vivo single-particle imaging of nuclear mRNA export in budding yeast demonstrates an essential role for Mex67p.
Smith C, Lari A, Derrer CP, Ouwehand A, Rossouw A, Huisman M, Dange T, Hopman M, Joseph A, Zenklusen D, Weis K, Grunwald D, Montpetit B. Smith C, et al. J Cell Biol. 2015 Dec 21;211(6):1121-30. doi: 10.1083/jcb.201503135. J Cell Biol. 2015. PMID: 26694837 Free PMC article. - Nuclear mRNA metabolism drives selective basket assembly on a subset of nuclear pore complexes in budding yeast.
Bensidoun P, Reiter T, Montpetit B, Zenklusen D, Oeffinger M. Bensidoun P, et al. Mol Cell. 2022 Oct 20;82(20):3856-3871.e6. doi: 10.1016/j.molcel.2022.09.019. Epub 2022 Oct 10. Mol Cell. 2022. PMID: 36220102 Free PMC article. - Into the basket and beyond: the journey of mRNA through the nuclear pore complex.
Ashkenazy-Titelman A, Shav-Tal Y, Kehlenbach RH. Ashkenazy-Titelman A, et al. Biochem J. 2020 Jan 17;477(1):23-44. doi: 10.1042/BCJ20190132. Biochem J. 2020. PMID: 31913454 Review. - Single particle imaging of mRNAs crossing the nuclear pore: Surfing on the edge.
Palazzo AF, Truong M. Palazzo AF, et al. Bioessays. 2016 Aug;38(8):744-50. doi: 10.1002/bies.201600038. Epub 2016 Jun 8. Bioessays. 2016. PMID: 27276446 Review. - Choosing the right exit: How functional plasticity of the nuclear pore drives selective and efficient mRNA export.
Bensidoun P, Zenklusen D, Oeffinger M. Bensidoun P, et al. Wiley Interdiscip Rev RNA. 2021 Nov;12(6):e1660. doi: 10.1002/wrna.1660. Epub 2021 May 2. Wiley Interdiscip Rev RNA. 2021. PMID: 33938148 Review.
Cited by
- The molecular architecture of the nuclear basket.
Singh D, Soni N, Hutchings J, Echeverria I, Shaikh F, Duquette M, Suslov S, Li Z, van Eeuwen T, Molloy K, Shi Y, Wang J, Guo Q, Chait BT, Fernandez-Martinez J, Rout MP, Sali A, Villa E. Singh D, et al. Cell. 2024 Sep 19;187(19):5267-5281.e13. doi: 10.1016/j.cell.2024.07.020. Epub 2024 Aug 9. Cell. 2024. PMID: 39127037 Free PMC article. - Altered RNA processing and export lead to retention of mRNAs near transcription sites and nuclear pore complexes or within the nucleolus.
Paul B, Montpetit B. Paul B, et al. Mol Biol Cell. 2016 Sep 1;27(17):2742-56. doi: 10.1091/mbc.E16-04-0244. Epub 2016 Jul 6. Mol Biol Cell. 2016. PMID: 27385342 Free PMC article. - In vivo single-particle imaging of nuclear mRNA export in budding yeast demonstrates an essential role for Mex67p.
Smith C, Lari A, Derrer CP, Ouwehand A, Rossouw A, Huisman M, Dange T, Hopman M, Joseph A, Zenklusen D, Weis K, Grunwald D, Montpetit B. Smith C, et al. J Cell Biol. 2015 Dec 21;211(6):1121-30. doi: 10.1083/jcb.201503135. J Cell Biol. 2015. PMID: 26694837 Free PMC article. - Quality control of mRNAs at the entry of the nuclear pore: Cooperation in a complex molecular system.
Soheilypour M, Mofrad MRK. Soheilypour M, et al. Nucleus. 2018 Jan 1;9(1):202-211. doi: 10.1080/19491034.2018.1439304. Nucleus. 2018. PMID: 29431587 Free PMC article. Review. - Structure of the pre-mRNA leakage 39-kDa protein reveals a single domain of integrated zf-C3HC and Rsm1 modules.
Hashimoto H, Ramirez DH, Lautier O, Pawlak N, Blobel G, Palancade B, Debler EW. Hashimoto H, et al. Sci Rep. 2022 Oct 21;12(1):17691. doi: 10.1038/s41598-022-22183-3. Sci Rep. 2022. PMID: 36271106 Free PMC article.
References
- Amberg D., Burke D., and Strathern J.. 2005. Methods in Yeast Genetics: A Cold Spring Harbor Laboratory Course Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. 230 pp.
- Cabal G.G., Genovesio A., Rodriguez-Navarro S., Zimmer C., Gadal O., Lesne A., Buc H., Feuerbach-Fournier F., Olivo-Marin J.C., Hurt E.C., and Nehrbass U.. 2006. SAGA interacting factors confine sub-diffusion of transcribed genes to the nuclear envelope. Nature. 441:770–773. 10.1038/nature04752 - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials