Swarm v2: highly-scalable and high-resolution amplicon clustering - PubMed (original) (raw)
Swarm v2: highly-scalable and high-resolution amplicon clustering
Frédéric Mahé et al. PeerJ. 2015.
Abstract
Previously we presented Swarm v1, a novel and open source amplicon clustering program that produced fine-scale molecular operational taxonomic units (OTUs), free of arbitrary global clustering thresholds and input-order dependency. Swarm v1 worked with an initial phase that used iterative single-linkage with a local clustering threshold (d), followed by a phase that used the internal abundance structures of clusters to break chained OTUs. Here we present Swarm v2, which has two important novel features: (1) a new algorithm for d = 1 that allows the computation time of the program to scale linearly with increasing amounts of data; and (2) the new fastidious option that reduces under-grouping by grafting low abundant OTUs (e.g., singletons and doubletons) onto larger ones. Swarm v2 also directly integrates the clustering and breaking phases, dereplicates sequencing reads with d = 0, outputs OTU representatives in fasta format, and plots individual OTUs as two-dimensional networks.
Keywords: Barcoding; Environmental diversity; Molecular operational taxonomic units.
Conflict of interest statement
The authors declare there are no competing interests.
Figures
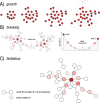
Figure 1. Schematic view of Swarm’s clustering and refinement approach.
(A) Swarm clusters amplicons iteratively by using a small user-chosen local threshold, d, allowing OTUs to grow to their natural limits, where no other amplicons can be added. (B) Swarm takes into account the abundance of each amplicon to produce higher resolution clusters, by not allowing the formation of amplicon chains. The darker the red, the higher the abundance. (C) The fastidious option avoids under-grouping (e.g., the production of small OTUs such as singletons and doubletons) by postulating the existence of virtual linking amplicons to graft smaller OTUs onto larger ones.
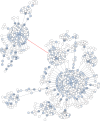
Figure 2. Graphical representation of an OTU produced by Swarm (breaking and grafting phases deactivated) when clustering the BioMarKs 18S rRNA V9 dataset (amplicons are appr. 129 bp in length).
Nodes represent amplicons. Node size, color and text annotations represent the abundance of each amplicon. Edges represent one difference (substitution, deletion or insertion); the length of the edges carries no information. The red-colored edge indicates where Swarm’s breaking phase cuts when it is not deactivated, resulting into two high abundant OTUs, each being assigned to a different genus of Collodaria (Radiolaria).
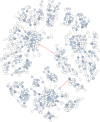
Figure 3. Graphical representation of an OTU produced by Swarm (breaking and grafting phases deactivated) when clustering the BioMarKs 18S rRNA V4 dataset (amplicons are appr. 380 bp in length).
Nodes represent amplicons. Node size, color and text annotations represent the abundance of each amplicon. Edges represent one difference (substitution, deletion or insertion); the length of the edges carries no information. The red-colored edges indicate where Swarm’s breaking phase cuts when it is not deactivated, resulting into three high abundant OTUs, each being assigned to a different taxa of Cnidaria (Metazoa).
Similar articles
- Swarm: robust and fast clustering method for amplicon-based studies.
Mahé F, Rognes T, Quince C, de Vargas C, Dunthorn M. Mahé F, et al. PeerJ. 2014 Sep 25;2:e593. doi: 10.7717/peerj.593. eCollection 2014. PeerJ. 2014. PMID: 25276506 Free PMC article. - Swarm v3: towards tera-scale amplicon clustering.
Mahé F, Czech L, Stamatakis A, Quince C, de Vargas C, Dunthorn M, Rognes T. Mahé F, et al. Bioinformatics. 2021 Dec 22;38(1):267-269. doi: 10.1093/bioinformatics/btab493. Bioinformatics. 2021. PMID: 34244702 Free PMC article. - Comparison of three clustering approaches for detecting novel environmental microbial diversity.
Forster D, Dunthorn M, Stoeck T, Mahé F. Forster D, et al. PeerJ. 2016 Feb 25;4:e1692. doi: 10.7717/peerj.1692. eCollection 2016. PeerJ. 2016. PMID: 26966652 Free PMC article. - GeFaST: An improved method for OTU assignment by generalising Swarm's fastidious clustering approach.
Müller R, Nebel ME. Müller R, et al. BMC Bioinformatics. 2018 Sep 12;19(1):321. doi: 10.1186/s12859-018-2349-1. BMC Bioinformatics. 2018. PMID: 30208838 Free PMC article. - Open-Source Sequence Clustering Methods Improve the State Of the Art.
Kopylova E, Navas-Molina JA, Mercier C, Xu ZZ, Mahé F, He Y, Zhou HW, Rognes T, Caporaso JG, Knight R. Kopylova E, et al. mSystems. 2016 Feb 9;1(1):e00003-15. doi: 10.1128/mSystems.00003-15. eCollection 2016 Jan-Feb. mSystems. 2016. PMID: 27822515 Free PMC article.
Cited by
- Terrestrial Inputs Shape Coastal Bacterial and Archaeal Communities in a High Arctic Fjord (Isfjorden, Svalbard).
Delpech LM, Vonnahme TR, McGovern M, Gradinger R, Præbel K, Poste AE. Delpech LM, et al. Front Microbiol. 2021 Feb 26;12:614634. doi: 10.3389/fmicb.2021.614634. eCollection 2021. Front Microbiol. 2021. PMID: 33717004 Free PMC article. - A Comparative Study of the Dynamics and Diversity of Bdellovibrio and Like Organisms in Lakes Annecy and Geneva.
Ezzedine JA, Scheifler M, Desdevises Y, Jacquet S. Ezzedine JA, et al. Microorganisms. 2022 Sep 30;10(10):1960. doi: 10.3390/microorganisms10101960. Microorganisms. 2022. PMID: 36296236 Free PMC article. - Serial cultures in invert emulsion and monophase systems for microbial community shaping and propagation.
Dijamentiuk A, Mangavel C, Gapp C, Elfassy A, Revol-Junelles AM, Borges F. Dijamentiuk A, et al. Microb Cell Fact. 2024 Feb 14;23(1):50. doi: 10.1186/s12934-024-02322-3. Microb Cell Fact. 2024. PMID: 38355580 Free PMC article. - Reintroducing mothur: 10 Years Later.
Schloss PD. Schloss PD. Appl Environ Microbiol. 2020 Jan 7;86(2):e02343-19. doi: 10.1128/AEM.02343-19. Print 2020 Jan 7. Appl Environ Microbiol. 2020. PMID: 31704678 Free PMC article. Review. - Facultative methanotrophs are abundant at terrestrial natural gas seeps.
Farhan Ul Haque M, Crombie AT, Ensminger SA, Baciu C, Murrell JC. Farhan Ul Haque M, et al. Microbiome. 2018 Jun 28;6(1):118. doi: 10.1186/s40168-018-0500-x. Microbiome. 2018. PMID: 29954460 Free PMC article.
References
- Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Pena AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R. QIIME allows analysis of high-throughput community sequencing data. Nature Methods. 2010;7(5):335–336. doi: 10.1038/nmeth.f.303. - DOI - PMC - PubMed
- De Vargas C, Audic S, Henry N, Decelle J, Mahé F, Logares R, Lara E, Berney C, Le Bescot N, Probert I, Carmichael M, Poulain J, Romac S, Colin S, Aury J-M, Bittner L, Chaffron S, Dunthorn M, Engelen S, Flegontova O, Guidi L, Horák A, Jaillon O, Lukeš J, Malviya S, Morard R, Mulot M, Scalco E, Siano R, Vincent F, Zingone A, Dimier C, Picheral M, Searson S, Kandels-Lewis S, Tara Oceans Coordinators. Acinas SG, Bork P, Bowler C, Gorsky G, Grimsley N, Hingamp P, Iudicone D, Not F, Ogata H, Pesant S, Raes J, Sieracki ME, Speich S, Stemman L, Sunagawa S, Weissenbach J, Wincker P, Karsenti E. Eukaryotic plankton diversity in the sunlit global ocean. Science. 2015;348(6237):1261605. doi: 10.1126/science.1261605. - DOI - PubMed
Grants and funding
FM and MD were supported by the Deutsche Forschungsgemeinschaft (grant #DU1319/1-1). CQ is funded by an EPSRC Career Acceleration Fellowship—EP/H003851/1. CdeV were supported by the EU EraNet BiodivErsA program BioMarKs (grant #2008-6530) and the French government “Investissements d’Avenir” project OCEANOMICS (ANR-11-BTBR-0008) and the EU FP7 program MicroB3 (contract number 287589). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
LinkOut - more resources
Full Text Sources
Other Literature Sources