Isolation and Characterization of a Novel Bat Coronavirus Closely Related to the Direct Progenitor of Severe Acute Respiratory Syndrome Coronavirus - PubMed (original) (raw)
. 2015 Dec 30;90(6):3253-6.
doi: 10.1128/JVI.02582-15.
Ben Hu 1, Bo Wang 1, Mei-Niang Wang 1, Qian Zhang 1, Wei Zhang 1, Li-Jun Wu 1, Xing-Yi Ge 1, Yun-Zhi Zhang 2, Peter Daszak 3, Lin-Fa Wang 4, Zheng-Li Shi 5
Affiliations
- PMID: 26719272
- PMCID: PMC4810638
- DOI: 10.1128/JVI.02582-15
Isolation and Characterization of a Novel Bat Coronavirus Closely Related to the Direct Progenitor of Severe Acute Respiratory Syndrome Coronavirus
Xing-Lou Yang et al. J Virol. 2015.
Abstract
We report the isolation and characterization of a novel bat coronavirus which is much closer to the severe acute respiratory syndrome coronavirus (SARS-CoV) in genomic sequence than others previously reported, particularly in its S gene. Cell entry and susceptibility studies indicated that this virus can use ACE2 as a receptor and infect animal and human cell lines. Our results provide further evidence of the bat origin of the SARS-CoV and highlight the likelihood of future bat coronavirus emergence in humans.
Copyright © 2016, American Society for Microbiology. All Rights Reserved.
Figures
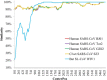
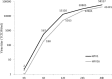
FIG 1
Similarity plot based on the nucleotide sequence of the S gene of bat SL-CoV WIV16. S genes of human/civet SARS-CoVs and bat SL-CoV WIV1 were used as reference sequences, with a window of 200 bp and a step size of 20 bp under the Kimura model. CenterPos, center position.
FIG 2
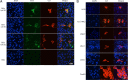
Receptor analysis (A) and susceptibility test (B) results for bat SL-CoV WIV16. (A) HeLa cells with and without the expression of ACE2. ACE2 expression was detected with goat anti-human ACE2 antibody, followed by fluorescein isothiocyanate (FITC)-conjugated donkey anti-goat IgG. Virus replication was detected with rabbit antibody against the SL-CoV Rp3 nucleocapsid protein, followed by cyanine 3 (Cy3)-conjugated mouse anti-rabbit IgG. Nuclei were stained with DAPI (4′,6-diamidino-2-phenylindole). The columns (from left to right) show staining of nuclei (blue), ACE2 expression (green), virus replication (red), and all three (merged triple-stain images). b, bat; c, civet; h, human. (B) Virus infection in A549, LLC-MK2, RSKT, PK15, H292, and Vero-E6 cells. The columns (from left to right) show staining of nuclei (blue), virus replication (red), and both nuclei and virus replication (merged double-stain images). A549 and H292, human lung cells; LLC-MK2, macaque kidney cells; RSKT, Chinese horseshoe bat kidney cells; PK15, pig kidney cells; VeroE6, African green monkey kidney cells.
FIG 3
One-step growth curve of bat SL-CoV WIV16 compared with that of WIV1. Vero E6 cells were infected with WIV16 or WIV1 at an MOI of 1. Supernatants were collected at 0, 12, 24, and 48 h postinfection. The viruses in the supernatant were determined by one-step reverse real-time PCR (n = 3). Virus RNA that had been extracted from a virus with a known titer was used to set up the standard curve. Error bars represent standard deviations. TCID50, 50% tissue culture infective doses.
Similar articles
- Evolutionary Arms Race between Virus and Host Drives Genetic Diversity in Bat Severe Acute Respiratory Syndrome-Related Coronavirus Spike Genes.
Guo H, Hu BJ, Yang XL, Zeng LP, Li B, Ouyang S, Shi ZL. Guo H, et al. J Virol. 2020 Sep 29;94(20):e00902-20. doi: 10.1128/JVI.00902-20. Print 2020 Sep 29. J Virol. 2020. PMID: 32699095 Free PMC article. - Isolation and characterization of a bat SARS-like coronavirus that uses the ACE2 receptor.
Ge XY, Li JL, Yang XL, Chmura AA, Zhu G, Epstein JH, Mazet JK, Hu B, Zhang W, Peng C, Zhang YJ, Luo CM, Tan B, Wang N, Zhu Y, Crameri G, Zhang SY, Wang LF, Daszak P, Shi ZL. Ge XY, et al. Nature. 2013 Nov 28;503(7477):535-8. doi: 10.1038/nature12711. Epub 2013 Oct 30. Nature. 2013. PMID: 24172901 Free PMC article. - Difference in receptor usage between severe acute respiratory syndrome (SARS) coronavirus and SARS-like coronavirus of bat origin.
Ren W, Qu X, Li W, Han Z, Yu M, Zhou P, Zhang SY, Wang LF, Deng H, Shi Z. Ren W, et al. J Virol. 2008 Feb;82(4):1899-907. doi: 10.1128/JVI.01085-07. Epub 2007 Dec 12. J Virol. 2008. PMID: 18077725 Free PMC article. - Angiotensin-converting enzyme 2: The old door for new severe acute respiratory syndrome coronavirus 2 infection.
Tan HW, Xu YM, Lau ATY. Tan HW, et al. Rev Med Virol. 2020 Sep;30(5):e2122. doi: 10.1002/rmv.2122. Epub 2020 Jun 30. Rev Med Virol. 2020. PMID: 32602627 Free PMC article. Review. - Molecular Basis of Pathogenesis of Coronaviruses: A Comparative Genomics Approach to Planetary Health to Prevent Zoonotic Outbreaks in the 21st Century.
Asrani P, Hasan GM, Sohal SS, Hassan MI. Asrani P, et al. OMICS. 2020 Nov;24(11):634-644. doi: 10.1089/omi.2020.0131. Epub 2020 Sep 16. OMICS. 2020. PMID: 32940573 Review.
Cited by
- Broad cross neutralizing antibodies against sarbecoviruses generated by SARS-CoV-2 infection and vaccination in humans.
Hu Y, Wu Q, Chang F, Yang J, Zhang X, Wang Q, Chen J, Teng S, Liu Y, Zheng X, Wang Y, Lu R, Pan D, Liu Z, Liu F, Xie T, Wu C, Tang Y, Tang F, Qian J, Chen H, Liu W, Li YP, Qu X. Hu Y, et al. NPJ Vaccines. 2024 Oct 22;9(1):195. doi: 10.1038/s41541-024-00997-8. NPJ Vaccines. 2024. PMID: 39438493 Free PMC article. - Molecular basis of convergent evolution of ACE2 receptor utilization among HKU5 coronaviruses.
Park YJ, Liu C, Lee J, Brown JT, Ma CB, Liu P, Xiong Q, Stewart C, Addetia A, Craig CJ, Tortorici MA, Alshukari A, Starr T, Yan H, Veesler D. Park YJ, et al. bioRxiv [Preprint]. 2024 Aug 28:2024.08.28.608351. doi: 10.1101/2024.08.28.608351. bioRxiv. 2024. PMID: 39253417 Free PMC article. Preprint. - Structural characteristics of BtKY72 RBD bound to bat ACE2 reveal multiple key residues affecting ACE2 usage of sarbecoviruses.
Su C, He J, Wang L, Hu Y, Cao J, Bai B, Qi J, Gao GF, Yang M, Wang Q. Su C, et al. mBio. 2024 Sep 11;15(9):e0140424. doi: 10.1128/mbio.01404-24. Epub 2024 Jul 31. mBio. 2024. PMID: 39082798 Free PMC article. - Binding affinity between coronavirus spike protein and human ACE2 receptor.
Shum MH, Lee Y, Tam L, Xia H, Chung OL, Guo Z, Lam TT. Shum MH, et al. Comput Struct Biotechnol J. 2024 Jan 17;23:759-770. doi: 10.1016/j.csbj.2024.01.009. eCollection 2024 Dec. Comput Struct Biotechnol J. 2024. PMID: 38304547 Free PMC article. Review.
References
- Guan Y, Zheng BJ, He YQ, Liu XL, Zhuang ZX, Cheung CL, Luo SW, Li PH, Zhang LJ, Guan YJ, Butt KM, Wong KL, Chan KW, Lim W, Shortridge KF, Yuen KY, Peiris JS, Poon LL. 2003. Isolation and characterization of viruses related to the SARS coronavirus from animals in southern China. Science 302:276–278. doi:10.1126/science.1087139. - DOI - PubMed
- Song HD, Tu CC, Zhang GW, Wang SY, Zheng K, Lei LC, Chen QX, Gao YW, Zhou HQ, Xiang H, Zheng HJ, Chern SW, Cheng F, Pan CM, Xuan H, Chen SJ, Luo HM, Zhou DH, Liu YF, He JF, Qin PZ, Li LH, Ren YQ, Liang WJ, Yu YD, Anderson L, Wang M, Xu RH, Wu XW, Zheng HY, Chen JD, Liang G, Gao Y, Liao M, Fang L, Jiang LY, Li H, Chen F, Di B, He LJ, Lin JY, Tong S, Kong X, Du L, Hao P, Tang H, Bernini A, Yu XJ, Spiga O, Guo ZM, Pan HY, He WZ, Manuguerra JC, Fontanet A, Danchin A, Niccolai N, Li YX, Wu CI, Zhao GP. 2005. Cross-host evolution of severe acute respiratory syndrome coronavirus in palm civet and human. Proc Natl Acad Sci U S A 102:2430–2435. doi:10.1073/pnas.0409608102. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous