In vivo gene editing in dystrophic mouse muscle and muscle stem cells - PubMed (original) (raw)
. 2016 Jan 22;351(6271):407-411.
doi: 10.1126/science.aad5177. Epub 2015 Dec 31.
Kexian Zhu # 1 3, Jason K W Cheng 1, Wei Leong Chew 2 4, Jeffrey J Widrick 5, Winston X Yan 6 7, Claire Maesner 1, Elizabeth Y Wu 1, Ru Xiao 8, F Ann Ran 6 7, Le Cong 6 7, Feng Zhang 6 7, Luk H Vandenberghe 8, George M Church 4, Amy J Wagers 1
Affiliations
- PMID: 26721686
- PMCID: PMC4924477
- DOI: 10.1126/science.aad5177
In vivo gene editing in dystrophic mouse muscle and muscle stem cells
Mohammadsharif Tabebordbar et al. Science. 2016.
Abstract
Frame-disrupting mutations in the DMD gene, encoding dystrophin, compromise myofiber integrity and drive muscle deterioration in Duchenne muscular dystrophy (DMD). Removing one or more exons from the mutated transcript can produce an in-frame mRNA and a truncated, but still functional, protein. In this study, we developed and tested a direct gene-editing approach to induce exon deletion and recover dystrophin expression in the mdx mouse model of DMD. Delivery by adeno-associated virus (AAV) of clustered regularly interspaced short palindromic repeats (CRISPR)-Cas9 endonucleases coupled with paired guide RNAs flanking the mutated Dmd exon23 resulted in excision of intervening DNA and restored the Dmd reading frame in myofibers, cardiomyocytes, and muscle stem cells after local or systemic delivery. AAV-Dmd CRISPR treatment partially recovered muscle functional deficiencies and generated a pool of endogenously corrected myogenic precursors in mdx mouse muscle.
Copyright © 2016, American Association for the Advancement of Science.
Figures
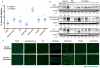
Figure 1. DYSTROPHIN expression in CRISPR-modified dystrophic satellite cells
(A) Detection of exon23 excision by genomic PCR in myotubes derived from satellite cells transfected with SpCas9 and Ai9 gRNAs (left lanes) or coupled Ai9-_Dmd_23 gRNAs (right lanes). Unedited genomic product, 1572bp; gene-edited product (red asterisk), 1189bp. M, molecular weight marker. (B) RT-PCR detection of exon23-deleted mRNA. Unedited RT-PCR product: 738bp; exon23-deleted product (blue asterisk): 525bp. (C) Western blot detecting DYSTROPHIN in myotubes derived from gene-edited satellite cells. A.U.: Arbitrary Unit, normalized to GAPDH (loading control). DYS: DYSTROPHIN, WT: wild type (D) DYSTROPHIN immunofluorescence in mdx muscles transplanted with satellite cells transfected ex vivo with SpCas9 + Ai9 gRNAs (top) or SpCas9 + Ai9-_Dmd_23 coupled gRNAs (bottom). For merge: Green: DYSTROPHIN; Red: tdTomato; Blue: DAPI (nuclei). Scale bar: 200 μm. See also Fig. S1I.
Figure 2. AAV-CRISPR enables in vivo excision of Dmd exon23 and restores DYSTROPHIN expression in adult dystrophic muscle
(A,B) Detection of exon23 excision in TA muscles from mdx;Ai9 mice injected intramuscularly with AAV-Ai9 CRISPR (left lanes) or AAV-Dmd_CRISPR (right lanes) by genomic PCR (A; unedited product 1012bp; exon-excised product 470bp) and RT-PCR (B). Asterisks mark gene-edited bands. M, molecular weight marker. (C) Western blot detecting DYSTROPHIN in muscles injected with AAV-Ai9 CRISPR (left) or AAV-Dmd CRISPR (right), with relative signal intensity determined by densitometry at bottom. A.U.: Arbitrary Unit, normalized to GAPDH. (D) Representative immunofluorescence images for DYSTROPHIN (green) and DAPI (blue) in_mdx;Ai9 muscles injected with AAV-Ai9 (left) or AAV-Dmd (right) CRISPR. Scale bar: 500 μm. (E,F) Muscle specific force (E) and decrease in force after eccentric damage (F) for wild type mice injected with vehicle (n=9), mdx;Ai9 mice injected with AAV-_Dmd_CRISPR in the right TA and vehicle in the left TA (n=12), mdx;Ai9 mice injected with AAV-Ai9 CRISPR in the right TA and vehicle in the left TA (n=12). *P<0.05, **P<0.01, n.s., not significant, One-Way ANOVA with Newman-Keuls multiple comparisons test.
Figure 3. Systemic dissemination of AAV-CRISPR targets Dmd exon23 and restores DYSTROPHIN in dystrophic cardiac and skeletal muscles
(A) Exon23-deleted transcripts in muscles quantified by Taqman. Data plotted for individual mice (n=7 receiving AAV-Dmd CRISPR (blue) and n=3 receiving AAV-Ai9 CRISPR (red)) and overlaid with mean ± SEM. (B) Western blots detecting DYSTROPHIN in the indicated muscles of mdx;Ai9 mice receiving systemic AAV-CRISPR. Right lanes correspond to muscles from 7 different mice injected intraperitoneally with AAV-Dmd CRISPR. Relative signal intensity, determined by densitometry, presented as A.U.: Arbitrary Unit normalized to GAPDH. (C) Representative immunofluorescence staining for DYSTROPHIN (green) in_mdx;Ai9_ mice injected with AAV-Ai9 (top) or AAV-Dmd(bottom) CRISPR. Blue: DAPI (nuclei). Scale bar: 200 μm.
Figure 4. Satellite cells in dystrophic muscles are transduced and targeted with systemically disseminated AAV-CRISPR
(A) Experimental design. (B) Percent of ZsGreen+ satellite cells expressing tdTomato+ after intraperitoneal injection of_Pax7-ZsGreen_+/−_;mdx;Ai9_ mice. Individual data points overlaid with mean ± SD; vehicle (n=3), AAV-Cre (n=4) AAV Ai9 CRISPR (n=5). (C) Representative immunofluorescence of myotubes differentiated from FACSorted satellite cells from mice injected intraperitoneally with vehicle, AAV-Cre, or AAV-Ai9 CRISPR. Green: Myosin heavy chain (MHC); Red: tdTomato. Blue: DAPI (nuclei) Scale bar: 200 μm. (D) Exon23-deleted DMD mRNA in satellite-cell derived myotubes from mice previously injected intraperitoneally with AAV-Dmd CRISPR (right lanes), compared to control AAV-Ai9 CRISPR (left lanes).
Comment in
- Genetic engineering: In vivo genome editing - growing in strength.
Koch L. Koch L. Nat Rev Genet. 2016 Mar;17(3):124. doi: 10.1038/nrg.2016.2. Epub 2016 Jan 19. Nat Rev Genet. 2016. PMID: 26781811 No abstract available. - CRISPR/Cas9 Flexes Its Muscles: In Vivo Somatic Gene Editing for Muscular Dystrophy.
VandenDriessche T, Chuah MK. VandenDriessche T, et al. Mol Ther. 2016 Mar;24(3):414-6. doi: 10.1038/mt.2016.29. Mol Ther. 2016. PMID: 26952918 Free PMC article. No abstract available.
Similar articles
- In vivo genome editing improves muscle function in a mouse model of Duchenne muscular dystrophy.
Nelson CE, Hakim CH, Ousterout DG, Thakore PI, Moreb EA, Castellanos Rivera RM, Madhavan S, Pan X, Ran FA, Yan WX, Asokan A, Zhang F, Duan D, Gersbach CA. Nelson CE, et al. Science. 2016 Jan 22;351(6271):403-7. doi: 10.1126/science.aad5143. Epub 2015 Dec 31. Science. 2016. PMID: 26721684 Free PMC article. - In Vivo Genome Editing Restores Dystrophin Expression and Cardiac Function in Dystrophic Mice.
El Refaey M, Xu L, Gao Y, Canan BD, Adesanya TMA, Warner SC, Akagi K, Symer DE, Mohler PJ, Ma J, Janssen PML, Han R. El Refaey M, et al. Circ Res. 2017 Sep 29;121(8):923-929. doi: 10.1161/CIRCRESAHA.117.310996. Epub 2017 Aug 8. Circ Res. 2017. PMID: 28790199 Free PMC article. - Muscle-specific CRISPR/Cas9 dystrophin gene editing ameliorates pathophysiology in a mouse model for Duchenne muscular dystrophy.
Bengtsson NE, Hall JK, Odom GL, Phelps MP, Andrus CR, Hawkins RD, Hauschka SD, Chamberlain JR, Chamberlain JS. Bengtsson NE, et al. Nat Commun. 2017 Feb 14;8:14454. doi: 10.1038/ncomms14454. Nat Commun. 2017. PMID: 28195574 Free PMC article. - The AAV-mediated and RNA-guided CRISPR/Cas9 system for gene therapy of DMD and BMD.
Wang JZ, Wu P, Shi ZM, Xu YL, Liu ZJ. Wang JZ, et al. Brain Dev. 2017 Aug;39(7):547-556. doi: 10.1016/j.braindev.2017.03.024. Epub 2017 Apr 5. Brain Dev. 2017. PMID: 28390761 Review. - Therapeutic Applications of CRISPR/Cas for Duchenne Muscular Dystrophy.
Wong TWY, Cohn RD. Wong TWY, et al. Curr Gene Ther. 2017;17(4):301-308. doi: 10.2174/1566523217666171121165046. Curr Gene Ther. 2017. PMID: 29173172 Review.
Cited by
- Interleukin-6 released by oral lichen planus myofibroblasts promotes angiogenesis.
Xu XH, Liu Y, Feng L, Yang YS, Liu SG, Guo W, Zhou HX, Li ZQ, Zhang L, Meng WX. Xu XH, et al. Exp Ther Med. 2021 Apr;21(4):291. doi: 10.3892/etm.2021.9722. Epub 2021 Jan 27. Exp Ther Med. 2021. PMID: 33717234 Free PMC article. - Human Autoinflammatory Diseases Mediated by NLRP3-, Pyrin-, NLRP1-, and NLRC4-Inflammasome Dysregulation Updates on Diagnosis, Treatment, and the Respective Roles of IL-1 and IL-18.
Alehashemi S, Goldbach-Mansky R. Alehashemi S, et al. Front Immunol. 2020 Aug 25;11:1840. doi: 10.3389/fimmu.2020.01840. eCollection 2020. Front Immunol. 2020. PMID: 32983099 Free PMC article. Review. - CRISPR/Cas9 with single guide RNA expression driven by small tRNA promoters showed reduced editing efficiency compared to a U6 promoter.
Wei Y, Qiu Y, Chen Y, Liu G, Zhang Y, Xu L, Ding Q. Wei Y, et al. RNA. 2017 Jan;23(1):1-5. doi: 10.1261/rna.057596.116. Epub 2016 Oct 14. RNA. 2017. PMID: 27742910 Free PMC article. - Genome-editing approaches and applications: a brief review on CRISPR technology and its role in cancer.
Siva N, Gupta S, Gupta A, Shukla JN, Malik B, Shukla N. Siva N, et al. 3 Biotech. 2021 Mar;11(3):146. doi: 10.1007/s13205-021-02680-4. Epub 2021 Feb 26. 3 Biotech. 2021. PMID: 33732568 Free PMC article. Review. - Emerging Medical Technologies and Their Use in Bionic Repair and Human Augmentation.
Manero A, Rivera V, Fu Q, Schwartzman JD, Prock-Gibbs H, Shah N, Gandhi D, White E, Crawford KE, Coathup MJ. Manero A, et al. Bioengineering (Basel). 2024 Jul 9;11(7):695. doi: 10.3390/bioengineering11070695. Bioengineering (Basel). 2024. PMID: 39061777 Free PMC article. Review.
References
- Koenig M, et al. Cell. 1987;50:509–517. - PubMed
- Tabebordbar M, Wang ET, Wagers AJ. Annu Rev Pathol. 2013;8:441–475. - PubMed
- Nakamura A, et al. Journal of clinical neuroscience : official journal of the Neurosurgical Society of Australasia. 2008;15:757–763. - PubMed
- Lu QL, et al. Nat Med. 2003;9:1009–1014. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- HHMI/Howard Hughes Medical Institute/United States
- T32 GM007753/GM/NIGMS NIH HHS/United States
- 5U01HL100402/HL/NHLBI NIH HHS/United States
- T32 GM008313/GM/NIGMS NIH HHS/United States
- 5R01DK097768-03/DK/NIDDK NIH HHS/United States
- PN2 EY018244/EY/NEI NIH HHS/United States
- 1DP2OD004345/OD/NIH HHS/United States
- 5DP1-MH100706/DP/NCCDPHP CDC HHS/United States
- 5PN2EY018244/EY/NEI NIH HHS/United States
- P50 HG005550/HG/NHGRI NIH HHS/United States
- DP2 OD004345/OD/NIH HHS/United States
- U01 HL100402/HL/NHLBI NIH HHS/United States
- DP1 MH100706/MH/NIMH NIH HHS/United States
- R01 MH110049/MH/NIMH NIH HHS/United States
- R01 DK097768/DK/NIDDK NIH HHS/United States
- T32 GM007598/GM/NIGMS NIH HHS/United States
- T2GM007753/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials