Characterization of bacterial isolates from the microbiota of mothers' breast milk and their infants - PubMed (original) (raw)
Characterization of bacterial isolates from the microbiota of mothers' breast milk and their infants
Kimberly Kozak et al. Gut Microbes. 2015.
Abstract
This investigation assessed the potential of isolating novel probiotics from mothers and their infants. A subset of 21 isolates among 126 unique bacteria from breast milk and infant stools from 15 mother-infant pairs were examined for simulated GI transit survival, adherence to Caco-2 cells, bacteriocin production, and lack of antibiotic resistance. Of the 21 selected isolates a Lactobacillus crispatus isolate and 3 Lactobacillus gasseri isolates demonstrated good profiles of in vitro GI transit tolerance and Caco-2 cell adherence. Bacteriocin production was observed only by L. gasseri and Enterococcus faecalis isolates. Antibiotic resistance was widespread, although not universal, among isolates from infants. Highly similar isolates (≥ 97% similarity by barcode match) of Bifidobacterium longum subsp. infantis (1 match), Lactobacillus fermentum (2 matches), Lactobacillus gasseri (6 matches), and Enterococcus faecalis (1 match) were isolated from 5 infant-mother pairs. Antibiotic resistance profiles between these isolate matches were similar, except in one case where the L. gasseri isolate from the infant exhibited resistance to erythromycin and tetracycline, not observed in matching mother isolate. In a second case, L. gasseri isolates differed in resistance to ampicillin, chloramphenicol and vancomycin between the mother and infant. In this study, gram positive bacteria isolated from mothers' breast milk as well as their infants exhibited diversity in GI transit survival and acid inhibition of pathogens, but demonstrated limited ability to produce bacteriocins. Mothers and their infants offer the potential for identification of probiotics; however, even in the early stages of development, healthy infants contain isolates with antibiotic resistance.
Keywords: antibiotic resistance; bacterial; breast milk; fecal; infant; microbiota; probiotics.
Figures
Figure 1.
Example of bacterial barcodes and isolate matching for Lactobacillus isolates from infant-mother pairs. Isolate IDs labeled with 126 are from infant stool and those labeled 124 are from the mother's breast milk.
Figure 2.
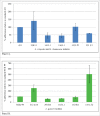
Caco-2 adherence of selected isolates of L. crispatus and L. rhamnosus (Panel A) and L. gasseri (Panel B) from infant stool and breast milk from infant-mother pairs. Reference strains: L. rhamnosus NCK 431 and L. gasseri NCK 99.
Figure 3.
Bacteriocin screening. Only those isolates, pathogens and indicator combinations for which a zone of inhibition was observed are shown. All other combinations of isolate and pathogens did not result in a zone of inhibition in this spot-overlay detection method. Panels A & B: L. gasseri 291R3 produces a protease (Proteinase K (K) and Pepsin (P)) sensitive bacteriocin against the indicator L. delbeueckii 235_,_ as indicated in Panel A. However, 291 R3's zone of inhibition against E. coli 0157:H7 is NOT sensitive to proteases. (Panel B). Further investigation was required to rule out acid production or an unusual bacteriocin structure. Panel C: Treatments: N- 10 N NaOH (3 μL) E- catalase (3 μL 10 mg/mL) Placing catalase in the zones had NO EFFECT; this ruled out hydrogen peroxide antimicrobial activity. But, placing 3 μL of 10 N NaOH inside the Zones of Inhibition (ZOI), diminished the antimicrobial activity by neutralizing the acid. Therefore, we conclude that the antimicrobial ZOIs against E. coli 0157:H7 are due to acid production. Panel D: The only isolate that produces a bacteriocin against a pathogen is E. faecalis 141 R4. This isolate produces a bacteriocin against Vancomycin Resistant Enterococcus (VRE) and is sensitive to Proteinase K, but not Pepsin (P).
Similar articles
- Bacteriocin production, antibiotic susceptibility and prevalence of haemolytic and gelatinase activity in faecal lactic acid bacteria isolated from healthy Ethiopian infants.
Birri DJ, Brede DA, Tessema GT, Nes IF. Birri DJ, et al. Microb Ecol. 2013 Feb;65(2):504-16. doi: 10.1007/s00248-012-0134-7. Epub 2012 Nov 27. Microb Ecol. 2013. PMID: 23184155 - Characterization of bacteriocin production in Lactobacillus spp. isolated from mother's milk.
Mohammadi F, Eshaghi M, Razavi S, Sarokhalil DD, Talebi M, Pourshafie MR. Mohammadi F, et al. Microb Pathog. 2018 May;118:242-246. doi: 10.1016/j.micpath.2018.03.020. Epub 2018 Mar 15. Microb Pathog. 2018. PMID: 29551436 - Probiotic potential of 3 Lactobacilli strains isolated from breast milk.
Martín R, Olivares M, Marín ML, Fernández L, Xaus J, Rodríguez JM. Martín R, et al. J Hum Lact. 2005 Feb;21(1):8-17; quiz 18-21, 41. doi: 10.1177/0890334404272393. J Hum Lact. 2005. PMID: 15681631 - Bifidobacterium longum subspecies infantis: champion colonizer of the infant gut.
Underwood MA, German JB, Lebrilla CB, Mills DA. Underwood MA, et al. Pediatr Res. 2015 Jan;77(1-2):229-35. doi: 10.1038/pr.2014.156. Epub 2014 Oct 10. Pediatr Res. 2015. PMID: 25303277 Free PMC article. Review. - Lactobacillus Bacteria in Breast Milk.
Łubiech K, Twarużek M. Łubiech K, et al. Nutrients. 2020 Dec 10;12(12):3783. doi: 10.3390/nu12123783. Nutrients. 2020. PMID: 33321792 Free PMC article. Review.
Cited by
- Assessment of some metabolic activities and potential probiotic properties of eight Enterococcus bacteria isolated from white cheese microbiota.
Hajikhani R, Onal Darilmaz D, Yuksekdag ZN, Beyatli Y. Hajikhani R, et al. Antonie Van Leeuwenhoek. 2021 Aug;114(8):1259-1274. doi: 10.1007/s10482-021-01599-3. Epub 2021 Jun 4. Antonie Van Leeuwenhoek. 2021. PMID: 34086120 - Gut Microbiota and Cardiovascular System: An Intricate Balance of Health and the Diseased State.
Bhat MA, Mishra AK, Tantray JA, Alatawi HA, Saeed M, Rahman S, Jan AT. Bhat MA, et al. Life (Basel). 2022 Nov 28;12(12):1986. doi: 10.3390/life12121986. Life (Basel). 2022. PMID: 36556351 Free PMC article. Review. - Intestinal Enterococcus abundance correlates inversely with excessive weight gain and increased plasma leptin in breastfed infants.
Laursen MF, Larsson MW, Lind MV, Larnkjær A, Mølgaard C, Michaelsen KF, Bahl MI, Licht TR. Laursen MF, et al. FEMS Microbiol Ecol. 2020 May 1;96(5):fiaa066. doi: 10.1093/femsec/fiaa066. FEMS Microbiol Ecol. 2020. PMID: 32275305 Free PMC article. - Infant gut microbiota modulation by human milk disaccharides in humanized microbiome mice.
Rubio-Del-Campo A, Gozalbo-Rovira R, Moya-Gonzálvez EM, Alberola J, Rodríguez-Díaz J, Yebra MJ. Rubio-Del-Campo A, et al. Gut Microbes. 2021 Jan-Dec;13(1):1-20. doi: 10.1080/19490976.2021.1914377. Gut Microbes. 2021. PMID: 33938391 Free PMC article.
References
- Hooper LV, Gordon JI. Commensal host-bacterial relationships in the gut. Science 2001; 292:1115-8; PMID:11352068; http://dx.doi.org/10.1126/science.1058709 - DOI - PubMed
- Berg RD. The indigenous gastrointestinal microflora. Trends Microbiol 1996; 4:430-5; PMID:8950812; http://dx.doi.org/10.1016/0966-842X(96)10057-3 - DOI - PubMed
- Makino H, Kushiro A, Ishikawa E, Muylaert D, Kubota H, Sakai T, Oishi K, Martin R, Ben Amor K, Oozeer R, et al. . Transmission of intestinal Bifidobacterium longum subsp. longum strains from mother to infant, determined by multilocus sequencing typing and amplified fragment length polymorphism. Appl Environ Microbiol 2011; 77:6788-93; PMID:21821739; http://dx.doi.org/10.1128/AEM.05346-11 - DOI - PMC - PubMed
- Albesharat R, Ehrmann MA, Korakli M, Yazaji S, Vogel RF. Phenotypic and genotypic analyses of lactic acid bacteria in local fermented food, breast milk and faeces of mothers and their babies. Syst Appl Microbiol 2011; 34:148-55; PMID:21300508; http://dx.doi.org/10.1016/j.syapm.2010.12.001 - DOI - PubMed
- Martin V, Maldonado-Barragan A, Moles L, Rodriguez-Banos M, Campo RD, Fernandez L, Rodríguez JM, Jiménez E. Sharing of bacterial strains between breast milk and infant feces. J Hum Lact 2012; 28:36-44; PMID:22267318; http://dx.doi.org/10.1177/0890334411424729 - DOI - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources