A cell cycle-dependent BRCA1-UHRF1 cascade regulates DNA double-strand break repair pathway choice - PubMed (original) (raw)
doi: 10.1038/ncomms10201.
Hailong Liu 2, Yali Chen 2, Xu Yang 2, Panfei Wang 2, Tongzheng Liu 3, Min Deng 3, Bo Qin 3, Cristina Correia 3, Seungbaek Lee 3, Jungjin Kim 3, Melanie Sparks 4, Asha A Nair 5, Debra L Evans 3, Krishna R Kalari 5, Pumin Zhang 6, Liewei Wang 7, Zhongsheng You 4, Scott H Kaufmann 3, Zhenkun Lou 2 3 7, Huadong Pei 2
Affiliations
- PMID: 26727879
- PMCID: PMC4728409
- DOI: 10.1038/ncomms10201
A cell cycle-dependent BRCA1-UHRF1 cascade regulates DNA double-strand break repair pathway choice
Haoxing Zhang et al. Nat Commun. 2016.
Abstract
BRCA1 is an important mediator of the DNA damage response, which promotes homologous recombination (HR) and antagonizes 53BP1-dependent non-homologous end joining in S/G2 phase. But how this is achieved remains unclear. Here, we report that the E3 ubiquitin ligase UHRF1 (Ubiquitin-like, with PHD and RING finger domains 1) directly participates in the interplay between BRCA1 and 53BP1. Mechanistically, UHRF1 is recruited to DNA double-strand breaks (DSBs) by BRCA1 in S phase, which requires the BRCT domain of BRCA1 and phosphorylated Ser674 of UHRF1. Subsequently, UHRF1 mediates K63-linked polyubiquitination of RIF1, and results in its dissociation from 53BP1 and DSBs thereby facilitating HR initiation. Thus, UHRF1 is a key regulator of DSB repair choice, which is separate from its role in heterochromatin formation and epigenetic regulator.
Conflict of interest statement
The authors declare no competing financial interests.
Figures
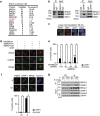
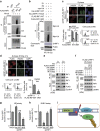
Figure 1. UHRF1 interacts with BRCA1 and is recruited to DNA damage sites by BRCA1 in S phase.
(a) Tandem affinity purification was performed using 293T cells stably expressing Flag-tagged BRCA1. The major hits from mass spectrometry analysis were shown in the table. (b) Reciprocal Co-IP between UHRF1 and BRCA1 in HeLa cells was performed. DBC1 was blotted as a negative control. (c) UHRF1 translocate to DSBs following DNA damage. U2OS cells were subjected to laser microirradiation to generate DSBs in a line pattern. Cells were then fixed after 30 min and immunostained with the indicated antibodies. Scale bar, 10 μm. (d,e) UHRF1 accumulation at DSBs requires BRCA1. U2OS cells were transfected with indicated constructs, then subjected to laser microirradiation to generate DSBs in a line pattern. Cells were then fixed and immunostained with the indicated antibodies (d) Representative micrographs (e) Quantification for (d), for each condition, 200 cells were counted. All the experiments were repeated three times biologically. Error bars represent the mean±s.d. of three biological triplicates. UHRF1 recruitment positive cell percentage compared with control group *P<0.05. NS: no siginficant difference. Scale bar, 10 μm. (f) UHRF1 recruitment to DSBs mainly occurs in S phase. U2OS cells were synchronized and subjected to laser microirradiation to generate DSBs in a line pattern. Cells were then fixed and immunostained with the indicated antibodies. Upper: representative micrographs; Lower: quantification. For each condition, 200 cells were counted. *P<0.05. NS: no siginficant difference. Scale bar, 10 μm. (g) The UHRF1–BRCA1 interaction is cell cycle regulated. HeLa cells were synchronized at G1 or S phase. Cell lysates were prepared and subjected to immunoprecipitation and immunoblot with the indicated antibodies.
Figure 2. BRCA1 recruits UHRF1 to DSBs through BRCT domain.
(a) GST-pull-down assay of UHRF1 using the indicated GST fusion proteins. (b) GST-pull-down assay of UHRF1 using wild-type BRCA1 BRCT domain or S1655A mutant. (c) UHRF1’s association with BRCA1 depends on phosphorylation. HEK 293T cells transfected with HA-UHRF1 were treated as indicated. The UHRF1–BRCA1 interaction was examined. (d) UHRF1 was phosphorylated by CDKs in S phase. HeLa cells were synchronized, and UHRF1 phosphorylation was examined with indicated antibodies. (e) CDKs inhibitor abolishes the interaction between UHRF1 and BRCA1 in S phase. HeLa cells were synchronized and treated with Roscovitine. UHRF1 phosphorylation was examined as indicated. (f) UHRF1 Ser674 was phosphorylated in S phase. HEK 293T cells transfected with the indicated constructs were synchronized at S phase, UHRF1 phosphorylation was examined. vec: empty vector. (g) UHRF1 is phosphorylated by CDK2/cyclin A. In vitro kinase assay was performed with CDK2/cyclin A using recombinant wild-type UHRF1 or UHRF1-S674A mutant as substrates. (h) Ser674 was important for UHRF1–BRCA1 interaction. HEK 293T cells stably expressing UHRF1 shRNA were transfected with indicated constructs. The BRCA1–UHRF1 interaction was examined following synchronization to S phase. (i) GST-BRCA1-BRCT fusion protein (17 nM to 2.5 μM) was passed over BIAcore chip (SA chip) surfaces immobilized with control peptide or phosphorylated S674 peptide. Resonance units were measured by BIAcore TP200. (j) Phosphorylated UHRF1 directly bind the BRCT domain of BRCA1. His-UHRF1 protein was subjected to CDK2/cyclin A phosphorylation or mock treatments in vitro, and resolved by SDS–PAGE. The binding of UHRF1 to the BRCT domain was analysed by Far-western blots. (k) Ser674 phosphorylation is important for UHRF1 accumulation at DSBs. U2OS cells were transfected with wild-type GFP-UHRF1 or S674A mutant. DSBs accumulation was monitored following laser microirradiation. For each sample, 200 cells were counted. Error bars represent the mean±s.d. of biological triplicates. Left: representative images; Right: quantification of positive signal. Error bars represent the mean±s.d. of biological triplicates. UHRF1 recruitment positive cell percentage compared with control group: *P<0.05. Scale bar, 10 μm. (l) Schematic model illustrating Figs 1–2.
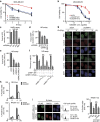
Figure 3. UHRF1 is involved in DNA repair through its E3 ligase activity.
(a) MDA-MB-231 cells stably expressing the indicated shRNAs were irradiated. Radiation sensitivity of cells was determined by colony formation assays. Data were presented as mean±s.d. of three biological triplicates. (b) MDA-MB-231 cells stably expressing indicated shRNAs were treated with increasing doses of PARP inhibitor (AZD2281). MTS assay was performed to determine the surviving fraction. Data presented as mean±s.d. of three biological triplicates. (c–e). HEK293 cells integrated with NHEJ or HR reporter were transfected with the indicated shRNAs and subjected to the NHEJ assay (c) or HR assay (d,e) as described in Methods. Data presented as mean±s.d. of three biological triplicates.. Positive cell percentage compared with control group. *P<0.05. NS: no siginficant difference. (**f**). UHRF1flox/hypo cells transfected with HR reporter were treated as indicated. HR assay were performed as described in Methods. Data presented as mean±s.d. of three biological triplicates. Positive cell percentage compared with control group. *_P_<0.05. NS: no siginficant difference. (**g**,**h**). UHRF1 regulates RPA and RAD51 IRIF. RPA and Rad51 foci formation were examined in synchronized HeLa cells stably expressing control or UHRF1 shRNA following irradiation (5 Gy). Cyclin A was stained as a cell cycle marker. (**g**) Representative micrographs. (**h**) Quantification data (foci>10 per cell). For each sample, randomly selected S phase cells (_n_=400) were counted. Data presented as mean±s.d. of three biological triplicates. Positive cell percentage compared with control group. *P<0.05. Scale bar, 10 μm. (**i**). HeLa cells stably expressing UHRF1 shRNA (targeting 3′-UTR) were reconstituted with the indicated constructs and synchronized in S phase. RAD51 foci were examined following irradiation (5 Gy 4 h). left: Representative micrographs. Right: Quantification of the positive cells (foci>10 per cell). For each sample, 400 randomly selected cells were counted. Data presented as mean±s.d. of three biological triplicates. Positive cell percentage compared with control group. *P<0.05. NS: no siginficant difference, Scale bar, 10 μm.
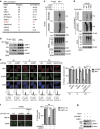
Figure 4. UHRF1 ubiquitinates RIF1.
(a) Tandem affinity purification of RIF1 was performed using antibody against endogenous RIF1. The major hits from mass spectrometry analysis were shown in the table. (b) Endogenous Co-IP between UHRF1 and RIF1 in HeLa cells was performed. 53BP1 and Znf 506 were blotted as positive and negative control respectively. (c) UHRF1 ubiquitinates RIF1 in vivo. HeLa cells stably expressing control or UHRF1 shRNA were irradiated (10 Gy), and RIF1 ubiquitination was then examined under denaturing conditions 1 h after IR (see Methods). (d) UHRF1-mediated K63-linked polyubiquitination of RIF1 in vivo. HEK 293T cells were transfected with the indicated constructs. RIF1 ubiquitination was examined as in c. (e) UHRF1 promotes removal of RIF1 from DSB sites in S phase. HeLa cells stably expressing the indicated shRNAs were synchronized and irradiated. RIF1 and H2AX foci formation were then examined. Left: Representative micrographs. Lower panels: representative cell cycle profiles of synchronized cells. Right: Quantitation of the positive cells (foci>10 per cell) as indicated. For each sample, 600 randomly selected cells were counted. Data presented as mean±s.d. of three biological triplicates. RIF1 foci Positive cell percentage compared with control group. *P<0.05. NS: no siginficant difference, Scale bar, 10 μm. (**f**) HeLa cells expressing the indicated shRNAs were irradiated and immunostained with antibodies as indicated. Left: representative micrographs. Right: quantification of BRCA1 and 53BP1 foci positive cells (foci>10 per cell). For each sample, randomly selected 500 cells were counted. Data presented as mean±s.d. of three biological triplicates. Positive cell percentage compared with control group. *P<0.05. NS: no siginficant difference, Scale bar, 10 μm. (g) Immunoblot with indicated antibodies for the cell lysates in f.
Figure 5. UHRF1 Ser674 phosphorylation is important for RIF1 ubiquitination.
(a) BRCA1 is required for the UHRF1–RIF1 interaction. HeLa cells expressing the indicated shRNAs were irradiated (10 Gy). The UHRF1–RIF1 interaction was examined 1 h following IR. (b) The S674A mutation abolishes UHRF1-mediated RIF1 ubiquitination. HEK 293T cells stably expressing UHRF1 shRNA were reconstituted with the indicated constructs. RIF1 ubiquitination was then examined in vivo. (c,d) Both UHRF1 S674 phosphorylation and UHRF1 E3 ligase activity regulate RIF1 accumulation at DSBs. HeLa cells stably expressing UHRF1 shRNA were reconstituted with the indicated constructs. RIF1 foci formation was then examined. (c) Representative micrographs. (d) Quantification of the positive cells. For each condition, 500 randomly selected cells were counted. Data presented as mean±s.d. of three biological triplicates. Positive cell percentage compared with WT group. *P<0.05. (e,f) UHRF1 depleted in HEK293 cells were reconstituted with the indicated constructs were subjected to HR assay (e) and NHEJ assay (f) as described in Methods. Data presented as mean±s.d. of three biological triplicates. Positive cell percentage compared with control group. *P<0.05. NS: no siginficant difference. (g) MDA-MB-231 cells stably expressing UHRF1 shRNA were reconstituted with the indicated constructs. Cell sensitivity to AZD2281 was examined by the MTS assay as in Fig. 3b. Data presented as the mean±s.d. of three biological triplicates.
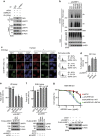
Figure 6. RIF1 ubiquitination by UHRF1 is important for its accumulation at DSB sites.
(a). Abrogation of RIF1 ubiquitination. HEK 293T cells stably expressing RIF1 shRNA were reconstituted with the indicated constructs. In vivo ubiquitination assay was performed as described in Methods. (b) UHRF1 ubiquitinates RIF1 in vitro. An in vitro ubiquitination assay was performed using recombinant UHRF1, ubiquitin, UBC13/Mms2 and UBE1 (See Methods). (c) Ubiquitination of RIF1 is required for its recruitment to the DSB sites. HeLa cells stably expressing RIF1 shRNA were reconstituted with the indicated constructs and synchronized. Flag-RIF1 focus formation was then examined following irradiation (5 Gy) as indicated. Upper: Representative micrographs. Lower: quantification of the positive cells (foci>10 per cell) and cell cycle profile. For each sample, 600 randomly selected cells were counted. Data presented as mean±s.d. of three biological triplicates. RIF1 foci Positive cell percentage compared between WT and 9KR group. *P<0.05. (**d**) Ubiquitination of RIF1 is required for RAD51 accumulation at DSBs. HeLa cells stably expressing RIF1 shRNA were reconstituted with the indicated constructs. Rad51 foci formation was examined 4 h following irradiation (5 Gy). Upper: Representative micrographs; Lower: Quantification of the positive cells (foci>10 per cell). For each sample, 600 randomly selected cells were counted. Data presented as mean±s.d. of three biological triplicates. *P<0.05. (e) HEK 293T cells stably expressing UHRF1 shRNA were transfected with the indicated constructs and synchronized at G1 or S phase, and 53BP1 and RIF1 interaction was examined. (f) HEK 293T cells stably expressing UHRF1 shRNA were reconstituted with the indicated constructs. 53BP1 and RIF1 interaction was examined following synchronization at S phase. (g,h) Ubiquitination of RIF1 is required for its function in DSB repair. RIF1 was depleted in HEK 293 cells integrated with HR or NHEJ reporter. Cells were reconstituted with the indicated constructs and subjected to HR assay (g) and NHEJ assay (h) as described in the Methods. Data presented as mean±s.d. of three biological triplicates. Positive cell percentage compared with control group. *P<0.05. NS: no siginficant difference. (i) Schematic model illustrating Figs 4.
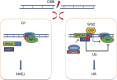
Figure 7
Model illustrating BRCA1-UHRF1 cascade regulating DNA repair choice.
Similar articles
- Regulation of repair pathway choice at two-ended DNA double-strand breaks.
Shibata A. Shibata A. Mutat Res. 2017 Oct;803-805:51-55. doi: 10.1016/j.mrfmmm.2017.07.011. Epub 2017 Jul 29. Mutat Res. 2017. PMID: 28781144 Review. - Functional crosstalk between DNA damage response proteins 53BP1 and BRCA1 regulates double strand break repair choice.
Bakr A, Köcher S, Volquardsen J, Reimer R, Borgmann K, Dikomey E, Rothkamm K, Mansour WY. Bakr A, et al. Radiother Oncol. 2016 May;119(2):276-81. doi: 10.1016/j.radonc.2015.11.001. Epub 2015 Nov 23. Radiother Oncol. 2016. PMID: 26615718 - A cell cycle-dependent regulatory circuit composed of 53BP1-RIF1 and BRCA1-CtIP controls DNA repair pathway choice.
Escribano-Díaz C, Orthwein A, Fradet-Turcotte A, Xing M, Young JT, Tkáč J, Cook MA, Rosebrock AP, Munro M, Canny MD, Xu D, Durocher D. Escribano-Díaz C, et al. Mol Cell. 2013 Mar 7;49(5):872-83. doi: 10.1016/j.molcel.2013.01.001. Epub 2013 Jan 17. Mol Cell. 2013. PMID: 23333306 - Methylation of UHRF1 by SET7 is essential for DNA double-strand break repair.
Hahm JY, Kim JY, Park JW, Kang JY, Kim KB, Kim SR, Cho H, Seo SB. Hahm JY, et al. Nucleic Acids Res. 2019 Jan 10;47(1):184-196. doi: 10.1093/nar/gky975. Nucleic Acids Res. 2019. PMID: 30357346 Free PMC article. - The influence of heterochromatin on DNA double strand break repair: Getting the strong, silent type to relax.
Goodarzi AA, Jeggo P, Lobrich M. Goodarzi AA, et al. DNA Repair (Amst). 2010 Dec 10;9(12):1273-82. doi: 10.1016/j.dnarep.2010.09.013. Epub 2010 Oct 30. DNA Repair (Amst). 2010. PMID: 21036673 Review.
Cited by
- PWWP2B promotes DNA end resection and homologous recombination.
Ju MK, Lee JR, Choi Y, Park SY, Sul HJ, Chung HJ, An S, Lee S, Jung E, Kim B, Choi BY, Kim BJ, Kim HS, Lim H, Kang HS, Soh JS, Myung K, Kim KC, Cho JW, Seo J, Kim TM, Lee JY, Kim Y, Kim H, Zang DY. Ju MK, et al. EMBO Rep. 2022 Jul 5;23(7):e53492. doi: 10.15252/embr.202153492. Epub 2022 May 18. EMBO Rep. 2022. PMID: 35582821 Free PMC article. - Interaction of the epigenetic integrator UHRF1 with the MYST domain of TIP60 inside the cell.
Ashraf W, Bronner C, Zaayter L, Ahmad T, Richert L, Alhosin M, Ibrahim A, Hamiche A, Mely Y, Mousli M. Ashraf W, et al. J Exp Clin Cancer Res. 2017 Dec 21;36(1):188. doi: 10.1186/s13046-017-0659-1. J Exp Clin Cancer Res. 2017. PMID: 29268763 Free PMC article. - How cells ensure correct repair of DNA double-strand breaks.
Her J, Bunting SF. Her J, et al. J Biol Chem. 2018 Jul 6;293(27):10502-10511. doi: 10.1074/jbc.TM118.000371. Epub 2018 Feb 5. J Biol Chem. 2018. PMID: 29414795 Free PMC article. Review. - Hemi-methylated DNA regulates DNA methylation inheritance through allosteric activation of H3 ubiquitylation by UHRF1.
Harrison JS, Cornett EM, Goldfarb D, DaRosa PA, Li ZM, Yan F, Dickson BM, Guo AH, Cantu DV, Kaustov L, Brown PJ, Arrowsmith CH, Erie DA, Major MB, Klevit RE, Krajewski K, Kuhlman B, Strahl BD, Rothbart SB. Harrison JS, et al. Elife. 2016 Sep 6;5:e17101. doi: 10.7554/eLife.17101. Elife. 2016. PMID: 27595565 Free PMC article. - Mouse Rif1 is a regulatory subunit of protein phosphatase 1 (PP1).
Sukackaite R, Cornacchia D, Jensen MR, Mas PJ, Blackledge M, Enervald E, Duan G, Auchynnikava T, Köhn M, Hart DJ, Buonomo SBC. Sukackaite R, et al. Sci Rep. 2017 May 18;7(1):2119. doi: 10.1038/s41598-017-01910-1. Sci Rep. 2017. PMID: 28522851 Free PMC article.
References
- Wyman C. & Kanaar R. DNA double-strand break repair: all's well that ends well. Annu. Rev. Genet. 40, 363–383 (2006). - PubMed
- Shrivastav M., De Haro L. P. & Nickoloff J. A. Regulation of DNA double-strand break repair pathway choice. Cell Res. 18, 134–147 (2008). - PubMed
- San Filippo J., Sung P. & Klein H. Mechanism of eukaryotic homologous recombination. Annu. Rev. Biochem. 77, 229–257 (2008). - PubMed
- Mattiroli F. et al. RNF168 ubiquitinates K13-15 on H2A/H2AX to drive DNA damage signaling. Cell 150, 1182–1195 (2012). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- NIH CA130996/CA/NCI NIH HHS/United States
- R01 CA122623/CA/NCI NIH HHS/United States
- P50 CA108961/CA/NCI NIH HHS/United States
- CA148940/CA/NCI NIH HHS/United States
- R01 CA116097/CA/NCI NIH HHS/United States
- R01 CA130996/CA/NCI NIH HHS/United States
- CA108961/CA/NCI NIH HHS/United States
- P50 CA116201/CA/NCI NIH HHS/United States
- R01 CA148940/CA/NCI NIH HHS/United States
- R01 CA203561/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous