The expression of heterologous MAM-7 in Lactobacillus rhamnosus reduces its intrinsic capacity to inhibit colonization of pathogen Vibrio parahaemolyticus in vitro - PubMed (original) (raw)
The expression of heterologous MAM-7 in Lactobacillus rhamnosus reduces its intrinsic capacity to inhibit colonization of pathogen Vibrio parahaemolyticus in vitro
Sebastian Beltran et al. Biol Res. 2016.
Abstract
Background: Vibrio parahaemolyticus (V. parahaemolyticus) is a Gram-negative, halophilic bacterium recognized as one of the most important foodborne pathogen. When ingested, V. parahaemolyticus causes a self-limiting illness (Vibriosis), characterized mainly by watery diarrhoea. Treatment is usually oral rehydration and/or antibiotics in complicated cases. Since 1996, the pathogenic and pandemic V. parahaemolyticus O3:K6 serotype has spread worldwide, increasing the reported number of vibriosis cases. Thus, the design of new strategies for pathogen control and illness prevention is necessary. Lactobacillus sp. grouped Gram positive innocuous bacteria, part of normal intestinal microbiota and usually used as oral vaccines for several diarrheic diseases. Recombinants strains of Lactobacillus (RL) expressing pathogen antigens can be used as part of an anti-adhesion strategy where RL block the pathogen union sites in host cells. Thus, we aimed to express MAM-7 V. parahaemolyticus adhesion protein in Lactobacillus sp. to generate an RL that prevents pathogen colonization.
Results: We cloned the MAM-7 gene from V. parahaemolyticus RIMD 2210633 in Lactobacillus expression vectors. Recombinant strains (Lactobacillus rhamnosus pSEC-MAM7 and L. rhamnosus pCWA-MAM7) adhered to CaCo-2 cells and competed with the pathogen. However, the L. rhamnosus wild type strain showed the best capacity to inhibit pathogen colonization in vitro. In addition, LDH-assay showed that recombinant strains were cytotoxic compared with the wild type isogenic strain.
Conclusions: MAM-7 expression in lactobacilli reduces the intrinsic inhibitory capacity of L. rhamnosus against V. parahaemolyticus.
Figures
Fig. 1
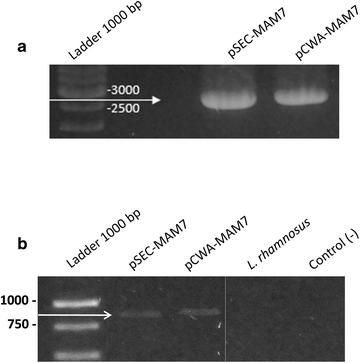
PCR and RT-PCR of MAM-7 from recombinants L. rhamnosus strains. a PCR of MAM-7 gene from L. rhamnosus pSEC-MAM-7 (pSEC-MAM7) and L. rhamnosus pCWA-MAM-7 (pCWA-MAM7) strains. b RT-PCR of MAM-7 transcript from L. rhamnosus pSEC-MAM-7 (pSEC-MAM7), L. rhamnosus pCWA-MAM-7 (pCWA-MAM7) and L. rhamnosus wild type (L. rhamnosus) strains
Fig. 2
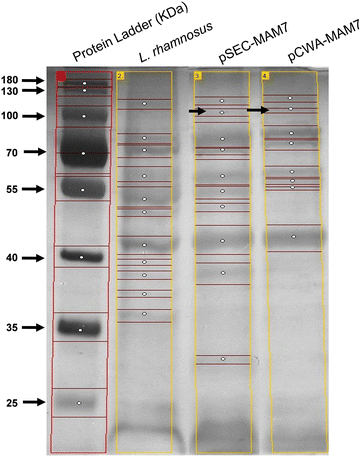
SDS-PAGE electrophoresis from recombinants L. rhamnsus strains. The figure shows the protein profile from L. rhamnosus pSEC-MAM-7 (pSEC-MAM7), L. rhamnosus pCWA-MAM-7 (pCWA-MAM7) and L. rhamnosus wild type (L. rhamnosus). Black arrows show a weakly band of approximately 100 kDa in recombinants strains. Bands of each lane were detected using GelAnalyzer and each white circle indicates a band
Fig. 3
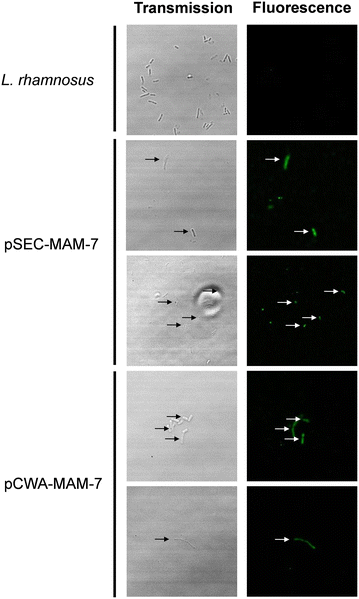
Expression of MAM-7 protein in L. rhamnosus. MAM-7 protein was detected using anti-Vibrio antibody and revealed with a secondary antibody alexa fluor® conjugated. The figures shows green fluorescence of alexa fluor® only in recombinant L. rhamnosus strains (L. rhamnosus pSEC-MAM-7 and L. rhamnosus pCWA-MAM-7). As a control, L. rhamnosus wild type was used
Fig. 4
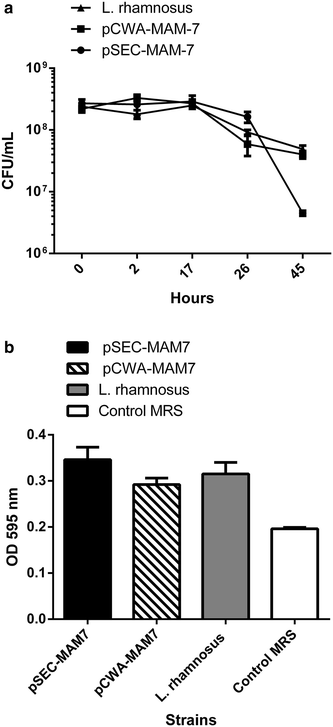
Effect of MAM-7 expression in L. rhamnosus in vitro growth. a Growth curve of L. rhamnosus pCWA-MAM-7 (black squares), L. rhamnosus pSEC-MAM-7 (black circles) recombinants strains and L. rhamnosus wild type strain (black triangle). b Biofilm formation of L. rhamnosus pSEC-MAM-7 (black column), L. rhamnosus pCWA-MAM-7 (striped column) recombinants strains and L. rhamnosus wild type strain (grey column). White column shows control with sterile MRS broth. The figure shows values expressed as the mean ± standard deviation of three full biological replicates, each time in technical triplicate
Fig. 5
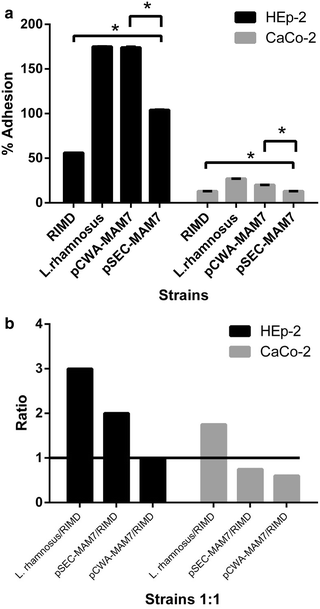
Adhesion and competition assays of V. parahaemolyticus and L. rhamnosus strains in epithelial cell lines. a Adhesion assay. The figure shows the adhesion percentage of L. rhamnosus strains and V. parahaemolyticus in epithelial cell line (HEp-2, black bars) and colon carcinoma cell line (CaCo-2, gray bars) monolayers. The strains used were V. parahaemolyticus RIMD 2210633 (RIMD), L. rhamnosus wild type (L. rhamnosus), _L. rhamnosus/_pCWA-MAM-7 (pCWA-MAM7) and _L. rhamnosus/_pSEC-MAM-7 (pSEC-MAM7). The results were expressed such as the percentage of adhesion at 3 h compared with initial inoculums of each strain at time zero. The figure shows values expressed as the mean ± standard deviation of three full biological replicates, each time in technical triplicate. *p < 0.001 (Student’s-Test). b Competition assay. The figure shows the results about the role of L. rhamnosus strains in the inhibition of V. parahaemolyticus from colonizing epithelial cell line (HEp-2, black bars) and colon carcinoma cell line (CaCo-2, gray bars) monolayers. The strains used were V. parahaemolyticus RIMD 2210633 (RIMD), L. rhamnosus wild type (L. rhamnosus), L. rhamnosus/pCWA-MAM-7 (pCWA-MAM7) and L. rhamnosus/pSEC-MAM-7 (pSEC-MAM7). Values expressed as a ration between the number of Lactobacillus strains UFC versus V. parahaemolyticus UFC after 3 h of monolayers 1:1 co-infection. The figure represents the result of three full biological replicates, each time in technical triplicate. The figure shows values expressed as the mean ± standard deviation of three full biological replicates, each time in technical triplicate. *p < 0.001 (Student’s-Test)
Fig. 6
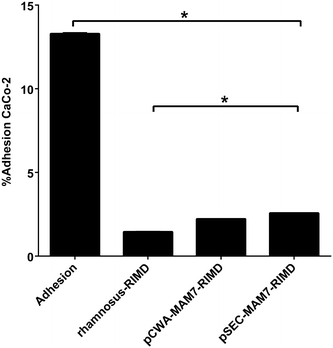
Competitive exclusion of L. rhamnosus to protect CaCo-2 cell line from V. parahaemolyticus colonization. The figure shows the results about the role of L. rhamnosus strains in the inhibition of V. parahaemolyticus from colonizing colon carcinoma cell line (CaCo-2) monolayers. The strains used were V. parahaemolyticus RIMD 2210633 (RIMD), L. rhamnosus wild type (L. rhamnosus), L. rhamnosus pCWA-MAM-7 (pCWA-MAM7) and L. rhamnosus pSEC-MAM-7 (pSEC-MAM7). Values are expressed as the adhesion percentage of V. parahaemolyticus after 3 h of L. rhamnosus wild type (L. rhamnosus– RIMD), pCWA-MAM-7 (pCWA-MAM7– RIMD) or pSEC-MAM-7 (pSEC-MAM7– RIMD) strains monolayer colonization. V. parahaemolyticus adhesion, without previous Lactobacillus treatment, is expressed as “Adhesion” bar. The figure shows values expressed as the mean ± standard deviation of three full biological replicates, each time in technical triplicate. *p < 0.001 (Student’s-Test)
Fig. 7
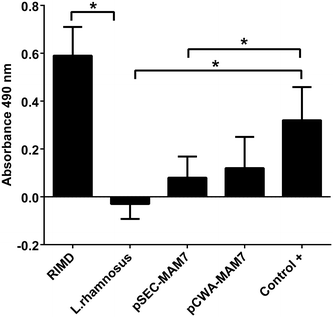
Cytotoxicity of V. parahaemolyticus and recombinants L. rhamnosus strains in CaCo-2 cell line. The figure shows LDH level release as absorbance at 490 nm of V. parahaemolyticus and Lactobacillus strains in colon carcinoma cell line (CaCo-2) monolayers. The strains used were V. parahaemolyticus RIMD 2210633 (RIMD), L. rhamnosus wild type (L. rhamnosus), L. rhamnosus pCWA-MAM-7 (pCWA-MAM7) and L. rhamnosus pSEC-MAM-7 (pSEC-MAM7). As a positive control (Control+), CaCo-2 were treated with Tritón ×100. The figure represents the result of three full biological replicates, each time in technical triplicate. *p < 0.001 (Student’s-Test)
Similar articles
- Alleviating effects of Lactobacillus strains on pathogenic Vibrio parahaemolyticus-induced intestinal fluid accumulation in the mouse model.
Yang ZQ, Jin CJ, Gao L, Fang WM, Gu RX, Qian JY, Jiao XA. Yang ZQ, et al. FEMS Microbiol Lett. 2013 Feb;339(1):30-8. doi: 10.1111/1574-6968.12050. Epub 2012 Dec 4. FEMS Microbiol Lett. 2013. PMID: 23210909 - PCR detection of a newly emerged pandemic Vibrio parahaemolyticus O3:K6 pathogen in pure cultures and seeded waters from the Gulf of Mexico.
Myers ML, Panicker G, Bej AK. Myers ML, et al. Appl Environ Microbiol. 2003 Apr;69(4):2194-200. doi: 10.1128/AEM.69.4.2194-2200.2003. Appl Environ Microbiol. 2003. PMID: 12676700 Free PMC article. - Pandemic Vibrio parahaemolyticus O3:K6 on the American continent.
Velazquez-Roman J, León-Sicairos N, de Jesus Hernández-Díaz L, Canizalez-Roman A. Velazquez-Roman J, et al. Front Cell Infect Microbiol. 2014 Jan 2;3:110. doi: 10.3389/fcimb.2013.00110. Front Cell Infect Microbiol. 2014. PMID: 24427744 Free PMC article. Review. - Epidemiology, pathogenesis, and prevention of foodborne Vibrio parahaemolyticus infections.
Yeung PS, Boor KJ. Yeung PS, et al. Foodborne Pathog Dis. 2004 Summer;1(2):74-88. doi: 10.1089/153531404323143594. Foodborne Pathog Dis. 2004. PMID: 15992266 Review.
Cited by
- Pediococcus pentosaceus LAB6- and Lactiplantibacillus plantarum LAB12-Derived Cell Free Supernatant Inhibited RhoA Activation and Reduced Amyloid-Β In Vitro.
Zaki RM, Ramasamy K, Ahmad Alwi NA, Mohd Yusoff R, Lim SM. Zaki RM, et al. Probiotics Antimicrob Proteins. 2024 Feb;16(1):62-75. doi: 10.1007/s12602-022-10009-7. Epub 2022 Nov 29. Probiotics Antimicrob Proteins. 2024. PMID: 36443559 - Lacticaseibacillus rhamnosus: A Suitable Candidate for the Construction of Novel Bioengineered Probiotic Strains for Targeted Pathogen Control.
Mathipa-Mdakane MG, Thantsha MS. Mathipa-Mdakane MG, et al. Foods. 2022 Mar 8;11(6):785. doi: 10.3390/foods11060785. Foods. 2022. PMID: 35327208 Free PMC article. Review. - Lactobacillus Mucosal Vaccine Vectors: Immune Responses against Bacterial and Viral Antigens.
LeCureux JS, Dean GA. LeCureux JS, et al. mSphere. 2018 May 16;3(3):e00061-18. doi: 10.1128/mSphere.00061-18. eCollection 2018 May-Jun. mSphere. 2018. PMID: 29769376 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources