Programmable polyproteams built using twin peptide superglues - PubMed (original) (raw)
Programmable polyproteams built using twin peptide superglues
Gianluca Veggiani et al. Proc Natl Acad Sci U S A. 2016.
Abstract
Programmed connection of amino acids or nucleotides into chains introduced a revolution in control of biological function. Reacting proteins together is more complex because of the number of reactive groups and delicate stability. Here we achieved sequence-programmed irreversible connection of protein units, forming polyprotein teams by sequential amidation and transamidation. SpyTag peptide is engineered to spontaneously form an isopeptide bond with SpyCatcher protein. By engineering the adhesin RrgA from Streptococcus pneumoniae, we developed the peptide SnoopTag, which formed a spontaneous isopeptide bond to its protein partner SnoopCatcher with >99% yield and no cross-reaction to SpyTag/SpyCatcher. Solid-phase attachment followed by sequential SpyTag or SnoopTag reaction between building-blocks enabled iterative extension. Linear, branched, and combinatorial polyproteins were synthesized, identifying optimal combinations of ligands against death receptors and growth factor receptors for cancer cell death signal activation. This simple and modular route to programmable "polyproteams" should enable exploration of a new area of biological space.
Keywords: antibody; nanobiotechnology; protein engineering; split protein; synthetic biology.
Conflict of interest statement
Conflict of interest statement: M.H. is an inventor on patent EP2534484, which applies to spontaneous isopeptide bond formation to a peptide tag, and United Kingdom Patent Application No. 1509782.7, which applies to iterative synthesis through isopeptide bonds.
Figures
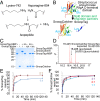
Fig. 1.
Establishing the covalently reactive peptide/protein pair SnoopTag and SnoopCatcher. (A) Spontaneous isopeptide bond formation between Lys and Asn, releasing ammonia. (B) Cartoon of splitting RrgA D4 domain [based on Protein Data Bank (PDB) ID code 2WW8] to make SnoopTag and SnoopCatcher. Reactive residues are in cyan. (C) SnoopTag-MBP reaction with SnoopCatcher, each at 10 µM, after 2 h at 25 °C analyzed by SDS/PAGE with Coomassie staining, alongside controls with Ala mutation of SnoopTag’s reactive Lys (KA) or SnoopCatcher’s reactive Asn (NA). (D) Isopeptide bond formation between SnoopTag peptide and SnoopCatcher shown by mass spectrometry. (E) Time course of SnoopTag reaction with a 1:1 or 2:1 ratio of SnoopCatcher to SnoopTag-MBP, tested as in C. (F) Time course of SnoopCatcher reaction with a 1:1, 2:1, or 4:1 ratio of SnoopTag-MBP to SnoopCatcher, tested as in C. Error bars are mean ±1 SD; n = 3. Some error bars are too small to be visible.
Fig. 2.
SnoopTag reaction is robust to conditions and orthogonal to SpyTag. (A) For pH dependence, 10 µM SnoopTag-MBP was incubated with 10 µM SnoopCatcher at the indicated pH for 15 min at 25 °C and analyzed by SDS/PAGE with Coomassie staining. (B) Temperature dependence of SnoopTag/SnoopCatcher reaction tested as in A. (C) SnoopTag/SnoopCatcher and SpyTag/SpyCatcher orthogonal reactivity, after incubation for 18 h at 25 °C, with each species at 10 µM, determined by SDS/PAGE with Coomassie staining. Error bars are all mean ± 1 SD; n = 3. Some error bars are too small to be visible.
Fig. S1.
SnoopTag and SnoopCatcher reaction conditions. (A) Point mutations to make SnoopCatcher. From the C-terminal domain of RrgA in cartoon format (based on PDB ID code 2WW8), residues mutated are shown in space-fill with carbons in cyan (G842 was changed to T and D848 to G). The Lys, Asn, and Glu involved in isopeptide bond formation are shown in stick format in yellow and the rest of SnoopTag colored magenta. (B) Sample gel to test for quantitative reaction of SnoopTag. Shown are 10 µM SnoopTag-MBP and 20 µM SnoopCatcher alone or mixed for 32 min (triplicate samples) before SDS/PAGE with Coomassie staining. (C) Sample gel to test for quantitative reaction of SnoopCatcher. Shown are 10 µM SnoopCatcher and 40 µM SnoopTag-MBP alone or mixed for the indicated times (triplicate samples) before SDS/PAGE with Coomassie staining. (D) Buffer dependence of SnoopTag/SnoopCatcher reaction. We incubated 10 µM SnoopTag-MBP with 10 µM SnoopCatcher at pH 8.0 for 15 min at 25 °C in the indicated buffer and analyzed the samples by SDS/PAGE with Coomassie staining. (E) TMAO dependence of SnoopTag/SnoopCatcher reaction tested as in D. Error bars are all mean ±1 SD; n = 3. Some error bars are too small to be visible.
Fig. S2.
SnoopCatcher reaction with SnoopTag was not reversible. (A) Reaction not reversed by competing SnoopTag. In lanes 1–8, SnoopCatcher, SnoopCatcher NA nonreactive control (mt), SnoopTag-MBP, and SnoopTag-Affibody (SnoopTag–AffiHER2–SpyTag) are shown alone (each at 10 µM) or after mixing for 6 h at 25 °C. In lane 9, 10 µM SnoopCatcher was reacted with 15 µM SnoopTag-MBP for 6 h, and then we added SnoopTag–AffiHER2–SpyTag to 130 µM for 16 h at 25 °C to look for SnoopTag–AffiHER2–SpyTag:SnoopCatcher formation. (B) Reaction not reversed by competing ammonia. We incubated SnoopTag-MBP with SnoopCatcher for 2 h in TBS with TMAO. We then added NH4Cl pH 9.0 to a 1 M final concentration to the indicated samples (+NH3) for 16 h (all at 25 °C) before SDS/PAGE with Coomassie staining. (The ammonium ion’s pKa is 9.2.)
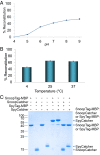
Fig. S3.
SpyTag and SpyCatcher reacted to a high degree of completion. (A) SpyTag with excess SpyCatcher. We incubated 20 µM SnoopTag–mEGFP–SpyTag with 40 µM SpyCatcher for 120 min at 25 °C before SDS/PAGE with Coomassie staining showing triplicate samples. (B) SpyCatcher with excess SpyTag. We incubated 10 µM SpyCatcher with 20 µM SpyTag-MBP for 32 min at 25 °C before SDS/PAGE with Coomassie staining showing triplicate samples.
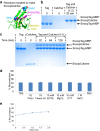
Fig. 3.
Solid-phase polyproteam synthesis. (A) Principle of polyproteam synthesis. Amylose resin is bound by modified MBP linked to SpyCatcher (MBPx-SpyCatcher). A protein of interest bearing SpyTag and SnoopTag is added to the resin and reacts with SpyCatcher on the growing chain (red line represents an isopeptide bond). SpyCatcher–SnoopCatcher is added and reacts with SnoopTag of the growing chain. Extension is continued by sequential additions of SnoopTag–X–SpyTag and SpyCatcher–SnoopCatcher. After extension is completed, the polyproteam is eluted from the resin with maltose. (B) Analysis of solid-phase polyproteam synthesis. Lanes 1–3 show MBPx-SpyCatcher, SpyCatcher–SnoopCatcher, and SnoopTag–AffiHER2–SpyTag in isolation. MBPx-SpyCatcher was bound to the amylose resin, and stepwise reaction with SnoopTag–AffiHER2–SpyTag and SpyCatcher–SnoopCatcher was carried out. After each stage, one aliquot of sample was eluted from the resin with maltose (lanes 4–13). Samples, without further purification, were analyzed by SDS/PAGE [both 4% and 10% (wt/vol) gels] with Coomassie staining.
Fig. S4.
Resin attachment for solid-phase polyprotein synthesis. (A) Location of MBP mutations, based on PDB ID code 1ANF. A312 was changed to V, I317 to V, and 172–173 and 175–176 were deleted. Mutated residues are shown in space-fill. The bound sugar part is shown in space-fill and is colored magenta. (B) Solid-phase polyprotein synthesis using avidin attachment. Lanes 1–3 show biotin-SpyCatcher, SnoopTag–AffiHER2–SpyTag, and SpyCatcher–SnoopCatcher in isolation. For solid-phase synthesis, biotin-SpyCatcher was bound to monomeric avidin resin. Sequential addition of SnoopTag–AffiHER2–SpyTag and SpyCatcher–SnoopCatcher led to chain formation up to a decamer. After each stage, sample was eluted from one aliquot of resin with biotin (lanes 4–13). Samples, without any further purification, were analyzed by SDS/PAGE [both 8% and 16% (wt/vol) gels] with Coomassie staining.
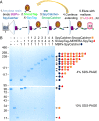
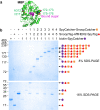
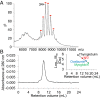
Fig. 4.
Biophysical analysis of polyproteams. (A) Electrospray ionization mass spectrometry to test identity of decamer polyproteam, MBPx-SpyCatcher:(SnoopTag–AffiHER2–SpyTag:SpyCatcher–SnoopCatcher)4:SnoopTag–AffiHER2–SpyTag. Red circles correspond to the decamer, with the charge state of the highest peak marked. (B) Size-exclusion chromatography of the same polyproteam. The Inset shows the molecular weight standards.
Fig. S5.
Stability of polyproteins. (A) Decamer thermostability. MBPx-SpyCatcher:(SnoopTag–AffiHER2–SpyTag:SpyCatcher–SnoopCatcher)4:SnoopTag–AffiHER2–SpyTag was incubated at the indicated temperature for 3 min, centrifuged to remove aggregates, and analyzed by SDS/PAGE with Coomassie staining. (B) Decamer time-dependent stability. Biotin-SpyCatcher:(SnoopTag–AffiHER2–SpyTag:SpyCatcher–SnoopCatcher)4:SnoopTag–AffiHER2–SpyTag was incubated at 25 °C for the indicated time, before boiling in SDS loading buffer and analysis by SDS/PAGE with Coomassie staining.
Fig. S6.
Different building blocks incorporated in polyproteams. (A) Synthesis of fluorescent polyproteins. MBPx-SpyCatcher was bound by amylose resin and then extended iteratively with SnoopTag–mEGFP–SpyTag and SpyCatcher–SnoopCatcher. Samples were eluted at each stage with maltose and analyzed by SDS/PAGE with Coomassie staining. (B) Synthesis of branched polyproteins, as for A, except using three tandemly linked affibodies (SnoopTag–SpyTag–AffiHER2 × 3).
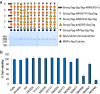
Fig. 5.
Combinatorial synthesis of polyproteams. (A) Chain synthesis with four anti-DR5 nanobodies and one affibody-based unit at various positions, analyzed by SDS/PAGE with Coomassie staining. (B) Polyprotein effect on viability of MDA–MB-231 cells after 40 h of incubation with each chain from A at 200 ng/mL, shown as a heat-map. (C) Viability of MDA–MB-231 cells at 40 h with titration of NNNNE and HHHHH control (N, anti-DR5 nanobody; E, AffiEGFR; H, AffiHER2 × 3). (D) Time course of cytotoxicity with 200 ng/mL NNNNE on MDA–MB-231 cells. (E) Caspase reporter activation in response to NNNNE, KillerTRAIL (a DR5 agonist) or HHHHH with or without Z-VAD-FMK pan-caspase inhibitor. (Error bars in each case are mean ± 1 SD; n = 3. Some error bars are too small to be visible.)
Fig. S7.
Effect of valency on polyproteam activation of cell death. (A) Synthesis of polyproteam homomultimers or heteromultimers with varying numbers of nanobodies and affibodies to either EGFR or Taq polymerase. Synthesis was analyzed by SDS/PAGE with Coomassie staining. (B) Polyproteam effect on viability of MDA–MB-231 after 40 h with each chain, analyzed by a resazurin assay with untreated cells set at 100% (mean ± 1 SD, n = 3). E, affibody to EGFR; H, triple affibody to HER2; I, affibody to IGF1R; N, nanobody to DR5; T, affibody to Taq polymerase.
Similar articles
- Structural analysis and optimization of the covalent association between SpyCatcher and a peptide Tag.
Li L, Fierer JO, Rapoport TA, Howarth M. Li L, et al. J Mol Biol. 2014 Jan 23;426(2):309-17. doi: 10.1016/j.jmb.2013.10.021. Epub 2013 Oct 23. J Mol Biol. 2014. PMID: 24161952 Free PMC article. - Dual Plug-and-Display Synthetic Assembly Using Orthogonal Reactive Proteins for Twin Antigen Immunization.
Brune KD, Buldun CM, Li Y, Taylor IJ, Brod F, Biswas S, Howarth M. Brune KD, et al. Bioconjug Chem. 2017 May 17;28(5):1544-1551. doi: 10.1021/acs.bioconjchem.7b00174. Epub 2017 May 5. Bioconjug Chem. 2017. PMID: 28437083 - Catching a SPY: Using the SpyCatcher-SpyTag and Related Systems for Labeling and Localizing Bacterial Proteins.
Hatlem D, Trunk T, Linke D, Leo JC. Hatlem D, et al. Int J Mol Sci. 2019 Apr 30;20(9):2129. doi: 10.3390/ijms20092129. Int J Mol Sci. 2019. PMID: 31052154 Free PMC article. Review. - Peptide tag forming a rapid covalent bond to a protein, through engineering a bacterial adhesin.
Zakeri B, Fierer JO, Celik E, Chittock EC, Schwarz-Linek U, Moy VT, Howarth M. Zakeri B, et al. Proc Natl Acad Sci U S A. 2012 Mar 20;109(12):E690-7. doi: 10.1073/pnas.1115485109. Epub 2012 Feb 24. Proc Natl Acad Sci U S A. 2012. PMID: 22366317 Free PMC article. - Secrets of a covalent interaction for biomaterials and biotechnology: SpyTag and SpyCatcher.
Reddington SC, Howarth M. Reddington SC, et al. Curr Opin Chem Biol. 2015 Dec;29:94-9. doi: 10.1016/j.cbpa.2015.10.002. Epub 2015 Oct 30. Curr Opin Chem Biol. 2015. PMID: 26517567 Review.
Cited by
- Glycosaminoglycan-Inspired Biomaterials for the Development of Bioactive Hydrogel Networks.
Neves MI, Araújo M, Moroni L, da Silva RMP, Barrias CC. Neves MI, et al. Molecules. 2020 Feb 21;25(4):978. doi: 10.3390/molecules25040978. Molecules. 2020. PMID: 32098281 Free PMC article. Review. - Greatest Hits-Innovative Technologies for High Throughput Identification of Bispecific Antibodies.
Hofmann T, Krah S, Sellmann C, Zielonka S, Doerner A. Hofmann T, et al. Int J Mol Sci. 2020 Sep 8;21(18):6551. doi: 10.3390/ijms21186551. Int J Mol Sci. 2020. PMID: 32911608 Free PMC article. Review. - Engineering a Novel Modular Adenoviral mRNA Delivery Platform Based on Tag/Catcher Bioconjugation.
Geng K, Rice-Boucher PJ, Kashentseva EA, Dmitriev IP, Lu ZH, Goedegebuure SP, Gillanders WE, Curiel DT. Geng K, et al. Viruses. 2023 Nov 20;15(11):2277. doi: 10.3390/v15112277. Viruses. 2023. PMID: 38005953 Free PMC article. - Protein Macrocyclization for Tertiary Structure Stabilization.
Haim A, Neubacher S, Grossmann TN. Haim A, et al. Chembiochem. 2021 Sep 2;22(17):2672-2679. doi: 10.1002/cbic.202100111. Epub 2021 Jun 21. Chembiochem. 2021. PMID: 34060202 Free PMC article. Review. - Injectable, photoresponsive hydrogels for delivering neuroprotective proteins enabled by metal-directed protein assembly.
Jiang B, Liu X, Yang C, Yang Z, Luo J, Kou S, Liu K, Sun F. Jiang B, et al. Sci Adv. 2020 Oct 9;6(41):eabc4824. doi: 10.1126/sciadv.abc4824. Print 2020 Oct. Sci Adv. 2020. PMID: 33036976 Free PMC article.
References
- Bachmann MF, Jennings GT. Vaccine delivery: A matter of size, geometry, kinetics and molecular patterns. Nat Rev Immunol. 2010;10(11):787–796. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials