Structure Determination of the Nuclear Pore Complex with Three-Dimensional Cryo electron Microscopy - PubMed (original) (raw)
Review
Structure Determination of the Nuclear Pore Complex with Three-Dimensional Cryo electron Microscopy
Alexander von Appen et al. J Mol Biol. 2016.
Abstract
Determining the structure of the nuclear pore complex (NPC) imposes an enormous challenge due to its size, intricate composition and membrane-embedded nature. In vertebrates, about 1000 protein building blocks assemble into a 110-MDa complex that fuses the inner and outer membranes of a cell's nucleus. Here, we review the recent progress in understanding the in situ architecture of the NPC with a specific focus on approaches using three-dimensional cryo electron microscopy. We discuss technological benefits and limitations and give an outlook toward obtaining a high-resolution structure of the NPC.
Keywords: cryoelectron microscopy; nuclear pore complex.
Copyright © 2016 The Authors. Published by Elsevier Ltd.. All rights reserved.
Figures
Graphical abstract
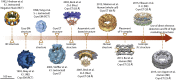
Fig. 1
3D structures of the NPC that were solved using EM. The timeline shows representative structures of the NPC drawn to scale. The model organism, the method used and the reported resolution are indicated when applicable. Images have been modified from the cited references (extracted: NPCs were extracted from membranes using heparin and/or detergent; Nuc: NPCs were embedded in the NE of intact nuclei; NE: NPCs were embedded into isolated NEs; whole cell: data were acquired on the intact cells).
Fig. 2
Overview of the NPC scaffold architecture and biological specimen used for its structure determination . (a) Cryoelectron micrograph of spread X.l. oocyte with high NPC density . (b) Three sequential x_–_y slices of 10 nm in thickness through a tomogram of a D.d. nucleus. Arrows indicate NPC in top view (left) and side view (right). Arrowheads show ribosomes decorating the outer nuclear membrane. (c) Isosurface rendering of the structure of the human NPC resolved to 33 Å seen from front (left), cut in half and tilted (right). Membranes are shown in brown. Dimensions of CR, IR and NR are indicated (adapted from Ref. [7]).
Fig. 3
The Y-complex assembles into two reticulated concentric rings within the nuclear and cytoplasmic rings of the NPC. (a) Negative staining structure of the human Y-complex at 33 Å resolution (top) that was localized by a systematic fitting approach within the tomographic map of the human NPC (32 Å resolution; middle) and the X.l. NPC structure (20 Å resolution; bottom). Images reproduced from Refs. , . (b) Crystal structure of the Y-complex vertex , (top) that was localized within the tomographic map of the human NPC resolved to 32 Å (bottom). (c) Fits of X-ray structures of sufficient size into the tomographic map of the human NPC resolved to 23 Å . The fit of the X-ray structure of the yeast vertex (top [53]) is shown in comparison to an independently obtained hybrid model of the entire Y-complex (bottom [51]).
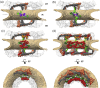
Fig. 4
Scaffold architecture of the NPC . The NPC is shown cut in half (a) four copies of the Y-complex per asymmetric unit of the NPC builds the scaffold of the cytoplasmic ring (CR) and the nucleoplasmic ring (NR). The outer copy is shown in orange; the inner copy is shown in gray. Multiple membrane contacts are apparent in the inner ring structure: density that contacts the outer leaf of the bilipid layer is shown in green and purple; apparent transmembrane domains are shown in blue. (b) Nup155 (green) appears to interact with the membrane at the points indicated in (a). (c) Six question mark densities (red) per asymmetric unit resembling the shapes of Nup205 or Nup188 localize to the CR, IR and NR. Another copy might reside only on the cytoplasmic site and is shown in orange. (d) Proteins of neighboring asymmetric units are shown as well.
Similar articles
- The Structure Inventory of the Nuclear Pore Complex.
Schwartz TU. Schwartz TU. J Mol Biol. 2016 May 22;428(10 Pt A):1986-2000. doi: 10.1016/j.jmb.2016.03.015. Epub 2016 Mar 22. J Mol Biol. 2016. PMID: 27016207 Free PMC article. Review. - Cryo-electron Microscopy Reveals the Structure of the Nuclear Pore Complex.
Tai L, Yin G, Sun F, Zhu Y. Tai L, et al. J Mol Biol. 2023 May 1;435(9):168051. doi: 10.1016/j.jmb.2023.168051. Epub 2023 Mar 17. J Mol Biol. 2023. PMID: 36933820 Review. - Perspective on the metazoan nuclear pore complex.
Maimon T, Medalia O. Maimon T, et al. Nucleus. 2010 Sep-Oct;1(5):383-6. doi: 10.4161/nucl.1.5.12332. Nucleus. 2010. PMID: 21326819 Free PMC article. - Structural analysis of a metazoan nuclear pore complex reveals a fused concentric ring architecture.
Frenkiel-Krispin D, Maco B, Aebi U, Medalia O. Frenkiel-Krispin D, et al. J Mol Biol. 2010 Jan 22;395(3):578-86. doi: 10.1016/j.jmb.2009.11.010. Epub 2009 Nov 11. J Mol Biol. 2010. PMID: 19913035 - The Structure of the Nuclear Pore Complex (An Update).
Lin DH, Hoelz A. Lin DH, et al. Annu Rev Biochem. 2019 Jun 20;88:725-783. doi: 10.1146/annurev-biochem-062917-011901. Epub 2019 Mar 18. Annu Rev Biochem. 2019. PMID: 30883195 Free PMC article. Review.
Cited by
- Nuclear Transport and Accumulation of Smad Proteins Studied by Single-Molecule Microscopy.
Li Y, Luo W, Yang W. Li Y, et al. Biophys J. 2018 May 8;114(9):2243-2251. doi: 10.1016/j.bpj.2018.03.018. Biophys J. 2018. PMID: 29742417 Free PMC article. - Host-HIV-1 Interactome: A Quest for Novel Therapeutic Intervention.
Shukla E, Chauhan R. Shukla E, et al. Cells. 2019 Sep 27;8(10):1155. doi: 10.3390/cells8101155. Cells. 2019. PMID: 31569640 Free PMC article. Review. - Cryo-EM structure of the inner ring from the Xenopus laevis nuclear pore complex.
Huang G, Zhan X, Zeng C, Liang K, Zhu X, Zhao Y, Wang P, Wang Q, Zhou Q, Tao Q, Liu M, Lei J, Yan C, Shi Y. Huang G, et al. Cell Res. 2022 May;32(5):451-460. doi: 10.1038/s41422-022-00633-x. Epub 2022 Mar 18. Cell Res. 2022. PMID: 35301439 Free PMC article. - A single-molecule localization microscopy method for tissues reveals nonrandom nuclear pore distribution in Drosophila.
Cheng J, Allgeyer ES, Richens JH, Dzafic E, Palandri A, Lewków B, Sirinakis G, St Johnston D. Cheng J, et al. J Cell Sci. 2021 Dec 15;134(24):jcs259570. doi: 10.1242/jcs.259570. Epub 2021 Dec 16. J Cell Sci. 2021. PMID: 34806753 Free PMC article. - A coarse-grained computational model of the nuclear pore complex predicts Phe-Gly nucleoporin dynamics.
Pulupa J, Rachh M, Tomasini MD, Mincer JS, Simon SM. Pulupa J, et al. J Gen Physiol. 2017 Oct 2;149(10):951-966. doi: 10.1085/jgp.201711769. Epub 2017 Sep 8. J Gen Physiol. 2017. PMID: 28887410 Free PMC article.
References
- Schwartz T.U. Modularity within the architecture of the nuclear pore complex. Curr. Opin. Struct. Biol. 2005;15(2):221–226. - PubMed
- Akey C.W. Structural plasticity of the nuclear pore complex. J. Mol. Biol. 1995;248(2):273–293. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources