Meta-connectomics: human brain network and connectivity meta-analyses - PubMed (original) (raw)
Review
Meta-connectomics: human brain network and connectivity meta-analyses
N A Crossley et al. Psychol Med. 2016 Apr.
Abstract
Abnormal brain connectivity or network dysfunction has been suggested as a paradigm to understand several psychiatric disorders. We here review the use of novel meta-analytic approaches in neuroscience that go beyond a summary description of existing results by applying network analysis methods to previously published studies and/or publicly accessible databases. We define this strategy of combining connectivity with other brain characteristics as 'meta-connectomics'. For example, we show how network analysis of task-based neuroimaging studies has been used to infer functional co-activation from primary data on regional activations. This approach has been able to relate cognition to functional network topology, demonstrating that the brain is composed of cognitively specialized functional subnetworks or modules, linked by a rich club of cognitively generalized regions that mediate many inter-modular connections. Another major application of meta-connectomics has been efforts to link meta-analytic maps of disorder-related abnormalities or MRI 'lesions' to the complex topology of the normative connectome. This work has highlighted the general importance of network hubs as hotspots for concentration of cortical grey-matter deficits in schizophrenia, Alzheimer's disease and other disorders. Finally, we show how by incorporating cellular and transcriptional data on individual nodes with network models of the connectome, studies have begun to elucidate the microscopic mechanisms underpinning the macroscopic organization of whole-brain networks. We argue that meta-connectomics is an exciting field, providing robust and integrative insights into brain organization that will likely play an important future role in consolidating network models of psychiatric disorders.
Keywords: Connectome; cytoarchitectonics; gene expression; graph theory; neuroimaging.
Conflict of interest statement
Declaration of Interest
ETB is employed half-time by the University of Cambridge and half-time by GlaxoSmithKline (GSK); he holds stock in GSK.
Figures
Fig. 1.
Functional connectivity and co-activation studies. (a) Functional connectivity is usually inferred from the correlation between two brain regions’ activities as registered in a resting-state functional magnetic resonance imaging (fMRI) scanning session. (b) Co-activation approaches infer functional connectivity from the frequency with which the two regions are reported to be active simultaneously during any task. As such, it is similar to traditional resting-state fMRI studies, in that it looks at the similarity in the activity between two regions. It differs in the time-frame used, where resting state looks at different volumes in one session, and co-activation looks at similarities across different published studies. The advantage of a co-activation approach is that one can infer the cognitive characteristics that elicit a functional connection between regions. In the figure shown, both regions were consistently co-activated during working memory tasks (squares), but not during visual tasks (diamond).
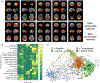
Fig. 2.
Cognition and the brain’s functional network organization. (a) Independent component analysis of co-activation data (right side of each individual panel) defines networks corresponding closely to the RSNs obtained from resting-state functional magnetic resonance imaging (left side of individual panels). (b) Analyses of the cognitive characteristics of the tasks showing co-activation in the above networks allowed characterization of the cognitive aspects of resting-state networks. Color scale shown is representing arbitrary units, in which each row has been normalized so that its mean value is one. ((a) and (b) reproduced from (Smith et al. 2009), with permission). (c) Graph theoretical analysis of functional co-activation data demonstrated a similar organization of cognitively specialized network modules, here depicted in groups with different colours, and in a spatial arrangement such that frequently co-activated nodes are located in close proximity to each other. Connecting these specialized modules is a more domain-general group of central and highly connected regions forming a rich club (nodes marked as squares) (reproduced from Crossley et al. 2013, with permission).
Fig. 3.
The network position of a node determines its role. (a) Social network analyses quantify the intuition that a central node or person (highlighted in red) with many connections would have more immediate access to many resources in the network, whether this is borrowing money or accessing information. However, if the most central agent loses contact with the rest of the network this can lead to disintegration of the network as a whole and the isolation of more peripheral agents. (b) Meta-analytic pooling of structural MRI studies from 26 different brain disorders demonstrated the existence of certain regions where abnormalities were consistently reported. (c) The probability of a brain region being “lesioned”, on average over all disorders, significantly increased as a logistic function of the degree of the corresponding regional node in the normative connectome. [Panels (b) and (c) from Crossley et al. 2014, with permission.]
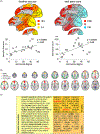
Fig. 4.
Relating cellular and molecular characteristics to macroscopic brain network organization. (a) Complexity of the neurons in layer III of the macaque, measured by the size of the dendritic tree and spine count, was correlated with the degree centrality of corresponding regional nodes of the macroscopic connectome (reproduced from Scholtens et al. 2014, with permission). (b) Human brain regions that are functionally connected to form resting state networks in fMRI data also have a high similarity in their transcription profiles. Organized correlated activity across the four networks described in (b) follows closely the transcription of the subset of genes shown in (c). Genes highlighted in bold code ion channels, and those in italic code neurotransmitters. All genes are grouped according to reliability of the data referred to as ‘splits’ in the original manuscript (see Richiardi et al. 2015 for details; reproduced with permission).
Similar articles
- The hubs of the human connectome are generally implicated in the anatomy of brain disorders.
Crossley NA, Mechelli A, Scott J, Carletti F, Fox PT, McGuire P, Bullmore ET. Crossley NA, et al. Brain. 2014 Aug;137(Pt 8):2382-95. doi: 10.1093/brain/awu132. Epub 2014 Jun 19. Brain. 2014. PMID: 25057133 Free PMC article. - Schizophrenia, neuroimaging and connectomics.
Fornito A, Zalesky A, Pantelis C, Bullmore ET. Fornito A, et al. Neuroimage. 2012 Oct 1;62(4):2296-314. doi: 10.1016/j.neuroimage.2011.12.090. Epub 2012 Feb 24. Neuroimage. 2012. PMID: 22387165 Review. - Identifying and Mapping Connectivity Patterns of Brain Network Hubs in Alzheimer's Disease.
Dai Z, Yan C, Li K, Wang Z, Wang J, Cao M, Lin Q, Shu N, Xia M, Bi Y, He Y. Dai Z, et al. Cereb Cortex. 2015 Oct;25(10):3723-42. doi: 10.1093/cercor/bhu246. Epub 2014 Oct 19. Cereb Cortex. 2015. PMID: 25331602 - Mapping migraine to a common brain network.
Burke MJ, Joutsa J, Cohen AL, Soussand L, Cooke D, Burstein R, Fox MD. Burke MJ, et al. Brain. 2020 Feb 1;143(2):541-553. doi: 10.1093/brain/awz405. Brain. 2020. PMID: 31919494 Free PMC article. - Depression, neuroimaging and connectomics: a selective overview.
Gong Q, He Y. Gong Q, et al. Biol Psychiatry. 2015 Feb 1;77(3):223-235. doi: 10.1016/j.biopsych.2014.08.009. Epub 2014 Aug 23. Biol Psychiatry. 2015. PMID: 25444171 Review.
Cited by
- The brain network hub degeneration in Alzheimer's disease.
Jin S, Wang J, He Y. Jin S, et al. Biophys Rep. 2024 Aug 31;10(4):213-229. doi: 10.52601/bpr.2024.230025. Biophys Rep. 2024. PMID: 39281195 Free PMC article. - Increasing variance of rich-club nodes distribution in early onset depression according to dynamic network.
Mai N, Wu Y, Zhong X, Chen B, Zhang M, Peng Q, Ning Y. Mai N, et al. Brain Imaging Behav. 2024 Jun;18(3):662-674. doi: 10.1007/s11682-023-00848-5. Epub 2024 Feb 13. Brain Imaging Behav. 2024. PMID: 38349505 - Structural networking of the developing brain: from maturation to neurosurgical implications.
De Benedictis A, Rossi-Espagnet MC, de Palma L, Sarubbo S, Marras CE. De Benedictis A, et al. Front Neuroanat. 2023 Nov 30;17:1242757. doi: 10.3389/fnana.2023.1242757. eCollection 2023. Front Neuroanat. 2023. PMID: 38099209 Free PMC article. Review. - Sex-dependent differences in connectivity patterns are related to episodic memory recall.
Spalek K, Coynel D, de Quervain D, Milnik A. Spalek K, et al. Hum Brain Mapp. 2023 Dec 1;44(17):5612-5623. doi: 10.1002/hbm.26465. Epub 2023 Aug 30. Hum Brain Mapp. 2023. PMID: 37647201 Free PMC article. - Editorial: Current state of the art of human brain white matter: From structural and functional connectivity to neurosurgical applications.
La Corte E, Ordóñez-Rubiano EG, Paiva WS, Johnson JM, Serrao G. La Corte E, et al. Front Neurol. 2022 Nov 22;13:1068212. doi: 10.3389/fneur.2022.1068212. eCollection 2022. Front Neurol. 2022. PMID: 36484018 Free PMC article. No abstract available.
References
- Bargmann CI, Marder E (2013). From the connectome to brain function. Nature Methods 10, 483–490. - PubMed
- Bullmore E, Sporns O (2009). Complex brain networks: graph theoretical analysis of structural and functional systems. Nature Reviews Neuroscience 10, 186–198. - PubMed
- Bullmore ET, Frangou S, Murray RM (1997). The dysplastic net hypothesis: an integration of developmental and dysconnectivity theories of schizophrenia. Schizophrenia Research 28, 143–156. - PubMed
Publication types
MeSH terms
Grants and funding
- 093875/Wellcome Trust/United Kingdom
- G1000183/MRC_/Medical Research Council/United Kingdom
- R01 MH074457/MH/NIMH NIH HHS/United States
- R01MH074457/MH/NIMH NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical