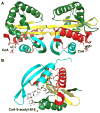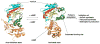Bacterial GCN5-Related N-Acetyltransferases: From Resistance to Regulation - PubMed (original) (raw)
Review
Bacterial GCN5-Related N-Acetyltransferases: From Resistance to Regulation
Lorenza Favrot et al. Biochemistry. 2016.
Abstract
The GCN5-related N-acetyltransferases family (GNAT) is an important family of proteins that includes more than 100000 members among eukaryotes and prokaryotes. Acetylation appears as a major regulatory post-translational modification and is as widespread as phosphorylation. N-Acetyltransferases transfer an acetyl group from acetyl-CoA to a large array of substrates, from small molecules such as aminoglycoside antibiotics to macromolecules. Acetylation of proteins can occur at two different positions, either at the amino-terminal end (αN-acetylation) or at the ε-amino group (εN-acetylation) of an internal lysine residue. GNAT members have been classified into different groups on the basis of their substrate specificity, and in spite of a very low primary sequence identity, GNAT proteins display a common and conserved fold. This Current Topic reviews the different classes of bacterial GNAT proteins, their functions, their structural characteristics, and their mechanism of action.
Conflict of interest statement
The authors declare no competing financial interest.
Figures
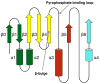
Figure 1
Topology scheme of the GNAT proteins. Starting at the N-terminal end, the secondary structure elements are colored dark green (_β_0, _β_1, _α_1, and _α_2), yellow (_β_2–_β_4), red (_α_3 and _β_5), and cyan (_α_4 and _β_6).
Figure 2
General structures of various aminoglycosides. The amino-cyclitol (2-deoxystreptamine) is colored blue. Arrows indicate the positions that are commonly modified by AACs. (A) 4,6-Disubstituted deoxystreptamine aminoglycoside. Examples cited in this review are butirosin, paromomycin, and neomycin. (B) 4,5-Disubstituted deoxystreptamine aminoglycoside. Position 2′ can be either an amino group (kanamycin B, gentamicin C1a and C1, etc.) or a hydroxyl group (kanamycin A, amikacin, etc.).
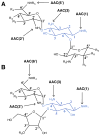
Figure 3
Structures of aminoglycoside _N_-acetyltransferases. The secondary structure elements are colored like the topology scheme presented in Figure 1. The substrates or products bound are rendered as sticks with white carbons. (A) Structure of S. marcescens AAC(3)-Ia (PDB entry 1BO4). (B) Structure of M. tuberculosis AAC(2′)-Ic in complex with CoA and kanamycin A (KAN) (PDB entry 1M4I). (C) Superimposition of the S. enterica AAC(6′)-Ib structure in complex with CoA and its variant AAC(6′)-Ib11 (PDB entries 2PRB and 2PR8, respectively). The _α_-helical flap (highlighted with an arrow in each structure) located above the aminoglycoside binding site is colored purple in the AAC(6′)-Ib structure and orange in the AAC(6′)-Ib11 structure.
Figure 4
Role and structure of MshD in mycothiol biosynthesis. (A) Biosynthetic pathway of mycothiol. The box highlights the reaction catalyzed by MshD. (B) Structure of M. tuberculosis MshD in complex with CoA and DAM (PDB entry 2C27). The secondary structure elements are colored like the topology scheme presented in Figure 1. The substrates or products bound are rendered as sticks with white carbons. (C) Superimposition of MshD in complex with CoA and DAM (PDB entry 2C27, gray) and MshD in complex with AcCoA (PDB entry 1OZP, beige). Upon substrate binding, the N-terminal domain rotates closer to the C-terminal domain to allow a narrower binding cavity for DAM as highlighted by the arrow.
Figure 5
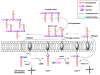
Scheme of peptidoglycan layer formation. Fem enzymes act at different stages of the peptide bridge formation depending on the bacterial species. In W. viridescens, FemX adds only the first amino acid. In S. aureus, FemX adds the first glycine residue whereas FemA adds the second and third glycine residues and FemB the last two glycine residues. For the sake of simplicity, only three of five amino acids in the peptide bridge are represented in the figure. After peptide bridge formation, the terminal Gly5 _α_-NH2 group of the peptide bridge is cross-linked to the penultimate amino acid of the neighboring pentapeptide stem by transpeptidation leading to the release of the terminal amino acid from the stem peptide.
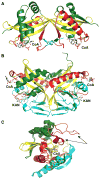
Figure 6
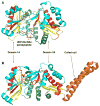
Structural comparison of W. viridescens FemX and S. aureus FemA. The GNAT secondary structure elements are colored like the topology scheme presented in Figure 1. The bisubstrate is rendered as sticks with white carbons. (A) The W. viridescens FemX consists of two similar GNAT domains, 1A and 1B (PDB entry 1P4N). (B) The S. aureus FemX structure also displays domains 1A and 1B and contains an additional coiled coil segment that is colored orange (PDB entry 1LRZ).
Figure 7
Structures of Rim proteins. The secondary structure elements are colored like the topology scheme presented in Figure 1. CoA and the bisubstrate are rendered as sticks with white carbons. (A) Structure of S. tphimurium LT2 RimL in complex with CoA (PDB entry 1S7N). (B) Structure of S. typhimurium LT2 RimI in complex with a bisubstrate CoA-S-acetyl-S181–6 (PDB entry 2CNM).
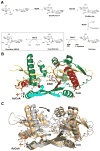
Figure 8
Activation of M. tuberculosis Pat via cAMP binding (left structure, PDB entry 4AVA; right structure, PDB entry 4AVC). The N-terminal regulatory domain (orange) is linked to C-terminal catalytic GNAT domain. Upon cAMP binding, the Pat protein undergoes a conformational rearrangement allowing protein–substrate binding in the catalytic domain active site. The GNAT secondary structure elements are colored like the topology scheme presented in Figure 1. The substrates and activators bound are rendered as sticks with white carbons.
Similar articles
- A bacterial acetyltransferase capable of regioselective N-acetylation of antibiotics and histones.
Vetting MW, Magnet S, Nieves E, Roderick SL, Blanchard JS. Vetting MW, et al. Chem Biol. 2004 Apr;11(4):565-73. doi: 10.1016/j.chembiol.2004.03.017. Chem Biol. 2004. PMID: 15123251 - Small-Molecule Acetylation by GCN5-Related _N_-Acetyltransferases in Bacteria.
Burckhardt RM, Escalante-Semerena JC. Burckhardt RM, et al. Microbiol Mol Biol Rev. 2020 Apr 15;84(2):e00090-19. doi: 10.1128/MMBR.00090-19. Print 2020 May 20. Microbiol Mol Biol Rev. 2020. PMID: 32295819 Free PMC article. Review. - Crystal structure of an aminoglycoside 6'-N-acetyltransferase: defining the GCN5-related N-acetyltransferase superfamily fold.
Wybenga-Groot LE, Draker K, Wright GD, Berghuis AM. Wybenga-Groot LE, et al. Structure. 1999 May;7(5):497-507. doi: 10.1016/s0969-2126(99)80066-5. Structure. 1999. PMID: 10378269 - Insights into the specificity of lysine acetyltransferases.
Tucker AC, Taylor KC, Rank KC, Rayment I, Escalante-Semerena JC. Tucker AC, et al. J Biol Chem. 2014 Dec 26;289(52):36249-62. doi: 10.1074/jbc.M114.613901. Epub 2014 Nov 7. J Biol Chem. 2014. PMID: 25381442 Free PMC article. - Structure and functions of the GNAT superfamily of acetyltransferases.
Vetting MW, S de Carvalho LP, Yu M, Hegde SS, Magnet S, Roderick SL, Blanchard JS. Vetting MW, et al. Arch Biochem Biophys. 2005 Jan 1;433(1):212-26. doi: 10.1016/j.abb.2004.09.003. Arch Biochem Biophys. 2005. PMID: 15581578 Review.
Cited by
- Acetylation of Lactate Dehydrogenase Negatively Regulates the Acidogenicity of Streptococcus mutans.
Ma Q, Pan Y, Chen Y, Yu S, Huang J, Liu Y, Gong T, Zhang Q, Sun Q, Zou J, Li Y. Ma Q, et al. mBio. 2022 Oct 26;13(5):e0201322. doi: 10.1128/mbio.02013-22. Epub 2022 Aug 31. mBio. 2022. PMID: 36043788 Free PMC article. - Insights into the Function of the _N_-Acetyltransferase SatA That Detoxifies Streptothricin in Bacillus subtilis and Bacillus anthracis.
Burckhardt RM, Escalante-Semerena JC. Burckhardt RM, et al. Appl Environ Microbiol. 2019 Mar 6;85(6):e03029-18. doi: 10.1128/AEM.03029-18. Print 2019 Mar 15. Appl Environ Microbiol. 2019. PMID: 30658980 Free PMC article. - Acetylation of glucosyltransferases regulates Streptococcus mutans biofilm formation and virulence.
Ma Q, Pan Y, Chen Y, Yu S, Huang J, Liu Y, Gong T, Zou J, Li Y. Ma Q, et al. PLoS Pathog. 2021 Dec 3;17(12):e1010134. doi: 10.1371/journal.ppat.1010134. eCollection 2021 Dec. PLoS Pathog. 2021. PMID: 34860858 Free PMC article. - The MinCDE Cell Division System Participates in the Regulation of Type III Secretion System (T3SS) Genes, Bacterial Virulence, and Motility in Xanthomonas oryzae pv. oryzae.
Yan Y, Wang Y, Yang X, Fang Y, Cheng G, Zou L, Chen G. Yan Y, et al. Microorganisms. 2022 Jul 31;10(8):1549. doi: 10.3390/microorganisms10081549. Microorganisms. 2022. PMID: 36013967 Free PMC article. - Escherichia coli ItaT is a type II toxin that inhibits translation by acetylating isoleucyl-tRNAIle.
Wilcox B, Osterman I, Serebryakova M, Lukyanov D, Komarova E, Gollan B, Morozova N, Wolf YI, Makarova KS, Helaine S, Sergiev P, Dubiley S, Borukhov S, Severinov K. Wilcox B, et al. Nucleic Acids Res. 2018 Sep 6;46(15):7873-7885. doi: 10.1093/nar/gky560. Nucleic Acids Res. 2018. PMID: 29931259 Free PMC article.
References
- Okamoto S, Suzuki Y. Chloramphenicol-, dihydrostreptomycin-, and kanamycin-inactivating enzymes from multiple drug-resistant Escherichia coli carrying episome ‘R’. Nature. 1965;208:1301–1303. - PubMed
- Brownell JE, Zhou J, Ranalli T, Kobayashi R, Edmondson DG, Roth SY, Allis CD. Tetrahymena histone acetyltransferase A: a homolog to yeast Gcn5p linking histone acetylation to gene activation. Cell. 1996;84:843–851. - PubMed
- Parthun MR, Widom J, Gottschling DE. The major cytoplasmic histone acetyltransferase in yeast: links to chromatin replication and histone metabolism. Cell. 1996;87:85–94. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical