Cell-Type-Specific Control of Brainstem Locomotor Circuits by Basal Ganglia - PubMed (original) (raw)
Cell-Type-Specific Control of Brainstem Locomotor Circuits by Basal Ganglia
Thomas K Roseberry et al. Cell. 2016.
Abstract
The basal ganglia (BG) are critical for adaptive motor control, but the circuit principles underlying their pathway-specific modulation of target regions are not well understood. Here, we dissect the mechanisms underlying BG direct and indirect pathway-mediated control of the mesencephalic locomotor region (MLR), a brainstem target of BG that is critical for locomotion. We optogenetically dissect the locomotor function of the three neurochemically distinct cell types within the MLR: glutamatergic, GABAergic, and cholinergic neurons. We find that the glutamatergic subpopulation encodes locomotor state and speed, is necessary and sufficient for locomotion, and is selectively innervated by BG. We further show activation and suppression, respectively, of MLR glutamatergic neurons by direct and indirect pathways, which is required for bidirectional control of locomotion by BG circuits. These findings provide a fundamental understanding of how BG can initiate or suppress a motor program through cell-type-specific regulation of neurons linked to specific actions.
Copyright © 2016 Elsevier Inc. All rights reserved.
Figures
Figure 1. Mapping and bidirectional BG regulation of the MLR
(A) Illustration of head-fixed trackball setup. (B) Schematic of stimulation within the MLR. (C) Population time course for mouse speed aligned to 20 Hz electrical stimulation (n = 7 mice). (D) MLR mapping using electrical stimulation. Green circles represent electrode placement at which stimulation elicited locomotion (> 5 cm/s) with short latency (< 2 s), red X’s represent electrode placement where no running was observed. SC, superior colliculus; IC, inferior colliculus; Cun, cuneiform nucleus; RRF, retrorubral field; MRN, mesencephalic reticular nucleus; RR, retrorubral nucleus; PPTg, pedunculopontine tegmentum; LL, lateral lemniscus; PB, parabrachial nucleus (n = 13 mice). (E) Schematic for stimulation of striatal dMSNs while recording activity in the MLR. (F) Example neuron excited during dMSN stimulation. Top, rasters of individual trials; bottom, peri-event time histogram (PSTH) of firing rate (purple line) and mouse speed (black line) aligned to onset of stimulation. (G) Histogram of AUCs for speed vs firing rate during a spontaneous locomotor session after dMSN stimulation. Black bars, neurons excited by dMSN stimulation (“dMSN exc”); gray bars, neurons unmodulated by dMSN stimulation (“dMSN nm”); open bars, neurons inhibited by dMSN stimulation (“dMSN inhib”), (n = 42 neurons from 4 mice). (H) Schematic for stimulation of striatal iMSNs while recording activity in the MLR. (I) Example neuron inhibited during iMSN stimulation as in (F). (J) Histogram of AUCs for speed vs firing rate during a spontaneous locomotor session after iMSN stimulation. Gray bars, neurons unmodulated by iMSN stimulation (“iMSN nm”); open bars, neurons inhibited by iMSN stimulation (“iMSN inhib”), (n = 26 neurons from 4 mice). All shaded regions, s.e.m.
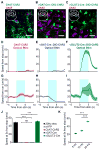
Figure 2. Distinct functions of MLR cell types
(A–C) Confocal images of coronal sections through the MLR of ChAT-ChR2 (A), vGAT-Cre::DIO-ChR2 (B) and vGLUT2-Cre::DIO-ChR2 (C) counter stained for ChAT, which demarcates the boundaries of the PPTg. Insets show location of image. (scale bar 25 μm) (D–I) Population time course for mouse speed aligned to 20 Hz optical stimulation from stationary (D–F) and running (G–I) states, in ChAT-ChR2 mice (D, G; n = 5 mice, 7 hemispheres), vGAT-Cre::DIO-ChR2 mice (E, H; n = 4 mice, 6 hemispheres) and vGLUT2-Cre::DIO-ChR2 mice (F, I; n = 6 mice, 7 hemispheres). Shaded regions, s.e.m. (J) Summary of population speed at 5 s after stimulation onset (*** p < 10-4 , Kruskal-Wallis one-way ANOVA, χ23,243 = 175.52, p < 10−10, with Dunn-Sidak post test). (K) Summary of speed at 5 s during graded stimulation of glutamatergic neurons (*** p < 10−4 , Kruskal-Wallis one-way ANOVA, χ22,167 = 175.52, p < 10−5, with Dunn-Sidak post test). All shaded regions, s.e.m. See also Figures S1 and S2.
Figure 3. Characterization of MLR glutamatergic neurons
(A) Schematic of optical tagging and recording setup in vGLUT2-Cre::DIO-ChR2 mice. (B) Recording sites for ChR2-positive (glutamatergic) neurons. (C) Top, light-evoked and spontaneous waveforms of an identified neuron. Bottom, PC1 vs PC2 for the neuron. Light gray dots, noise; dark gray dots, spontaneous spikes; blue dots, light-evoked spikes. (D) Raster (top) and PSTH (bottom) for a light-reactive neuron showing 3 ms latency aligned to laser onset. (E) Smoothed firing rate of the neuron identified in (C) and (D) (green line, left axis) plotted with the speed of the mouse (black line, right axis). (F) Histogram of AUC’s for all recorded neurons during spontaneous locomotion. Green bars, identified glutamatergic neurons (all significantly encoded the stationary or locomotor state); filled grey bars, unidentified neurons recorded in the MLR from a separate cohort that significantly encoded the stationary or running state; open grey bars represent neurons which did not significantly encode either state. Arrows, means; ** p < 0.001, Wilcoxon rank sum. (G) Histogram of R2 values for the glutamatergic neurons that predicted locomotion in (F) (n = 14). Light green bars, speed correlated neurons; dark green bars, neurons not correlated with speed. (H) Population z-scored firing rate of identified glutamatergic neurons (green trace) aligned to onset of locomotion. Black trace, speed. Inset, individual example firing rate traces (light green) and average (dark green) aligned to starts. (I) Probability of a start within one second given z-scored firing rate. Each point represents one binned data point (0.1 sd bins) from one neuron. All shaded regions, s.e.m. See also Figure S3.
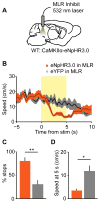
Figure 4. Inhibition of MLR glutamatergic neurons impedes running
(A) Experimental schematic for inhibiting glutamatergic neurons in the MLR. (B) Speed aligned to laser onset, or with no stimulation (eNpHR3.0 in MLR, orange line, n = 4 mice; eYFP in MLR, black line, n = 6 mice). (C) Number of stops during laser inhibition for each group. (D) Speed summary data at 5 seconds after onset of laser inhibition. * p < 0.05, ** p < 0.01, Wilcoxon rank sum. Shaded regions, s.e.m.
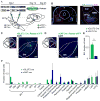
Figure 5. Brain-wide mapping of inputs to MLR glutamatergic and GABAergic neurons
(A) Schematic and time course of experiment. (B) Left, coronal section through the MLR showing TVA-mCherry labeling (blue) around the PPTg (labelled with ChAT staining, pink) and center-of-mass of TVA and RG injection sites. (red dots, vGAT-Cre::TVA+RG; green X’s, vGLUT2-Cre::TVA+RG). Right, close-up of infected cells. Scale bars, 500 μm. (C–D) Examples of eYFP expression in the SNr in a vGLUT2-Cre::Rabies-eYFP (C) and vGAT-Cre::Rabies-eYFP (D). Scale bars, 200 μm. (E) Quantification of labelled cell counts from all BG nuclei in vGLUT2-Cre::Rabies-eYFP (green bars) and vGAT-Cre::Rabies-eYFP mice (blue bars). ** p < 0.01, Wilcoxon rank sum. (F) Quantification of ipsilateral inputs to MLR glutamatergic (green) and GABAergic (blue) neurons. SNr, substantia nigra pars reticulate; SNc, substantia nigra pars compacta; DMS, dorsomedial striatum; VS, ventral striatum; STN, subthalamic nucleus; GPe, external globus pallidus; EP, entopeduncular nucleus; VTA, ventral tegmental area; CeA, central amygdalar nucleus; LDTg, laterodorsal tegmental nucleus. Error bars, s.e.m.
Figure 6. MLR glutamatergic neurons are necessary and sufficient to reverse the effects of BG stimulation
(A) Schematic for recording MLR glutamatergic neurons, while activating dMSNs in striatum. (B) Left, Z-scored firing rate of identified glutamatergic neurons (green line, left axis) and speed (black line, right axis) aligned to onset of 5 second unilateral dMSN stimulation from stop. Right, fractions of excited (excite), inhibited (inhib), or non-modulated (nm) units in unidentified recordings (un-IDed, left) and in identified glutamatergic cells (IDed, right). (identified: 25 excited, 0 inhibited, 1 non-modulated from 4 mice; unidentified: 22 excited, 11 inhibited, 9 non-modulated from 4 mice; ** p < 0.001, χ2 test). (C) Schematic for stimulating dMSNs in striatum, while inhibiting MLR glutamatergic neurons. (D) Left, Speed aligned to unilateral dMSN stimulation. Orange line, trials in which green light was turned on in the MLR 5 seconds after dMSN stimulation; black line, interleaved trials in which green light was omitted; purple line, no stimulation trials when mouse is stopped. Right, Speed 10 seconds after onset of dMSN stimulation (*** p < 10−4, Kruskal-Wallis one-way ANOVA, χ23,136 = 92.35, p < 10−10, with Dunn-Sidak post test). (E) Schematic for recording identified MLR glutamatergic neurons, while activating iMSNs in striatum. (F) Left, Z-scored firing rate of glutamatergic neurons (green line, left axis) and speed (black line, right axis) aligned to onset of 5 second bilateral iMSN stimulation from running. Right, summaries for the number of inhibited (inhib) or non-modulated (nm) units in unidentified (unIDed, left) recordings and identified (IDed, right) glutamatergic neurons (identified: 25 inhibited, 2 unmodulated from 4 mice; unidentified: 18 inhibited, 8 unmodulated from 3 mice; p = 0.09, χ2 test). (G) Schematic for stimulating iMSNs in striatum while stimulating glutamatergic cells in the MLR. (H) Left, time course of mouse speed aligned to bilateral iMSN stimulation. Red line, trials in which blue light was turned on in the MLR at 20 Hz 5 seconds after iMSN stimulation; black line, interleaved trials in which green light was omitted; purple line, no stimulation trials when mouse is running. Right, summary of mouse speed 10 seconds after onset of iMSN stimulation (*** p < 10−4 , Kruskal-Wallis one-way ANOVA, χ23,105 = 75.06, p < 10−10, with Dunn-Sidak post test). Shaded regions, s.e.m. See also Figures S5 and S6.
Comment in
- Motor Control: Illuminating an Enigmatic Midbrain Locomotor Center.
Esposito MS, Arber S. Esposito MS, et al. Curr Biol. 2016 Apr 4;26(7):R291-3. doi: 10.1016/j.cub.2016.02.043. Curr Biol. 2016. PMID: 27046818 - Are the basal ganglia actually controlling movement or quite the opposite?
Trigo-Damas I, Gonzalez Del Rey NL. Trigo-Damas I, et al. Mov Disord. 2016 Sep;31(9):1339. doi: 10.1002/mds.26680. Epub 2016 Jun 6. Mov Disord. 2016. PMID: 27270789 No abstract available.
Similar articles
- The Basal Ganglia and Mesencephalic Locomotor Region Connectivity Matrix.
Morgenstern NA, Esposito MS. Morgenstern NA, et al. Curr Neuropharmacol. 2024;22(9):1454-1472. doi: 10.2174/1570159X21666230809112840. Curr Neuropharmacol. 2024. PMID: 37559244 Free PMC article. Review. - Control of basal ganglia output by direct and indirect pathway projection neurons.
Freeze BS, Kravitz AV, Hammack N, Berke JD, Kreitzer AC. Freeze BS, et al. J Neurosci. 2013 Nov 20;33(47):18531-9. doi: 10.1523/JNEUROSCI.1278-13.2013. J Neurosci. 2013. PMID: 24259575 Free PMC article. - Functional diversity for body actions in the mesencephalic locomotor region.
Ferreira-Pinto MJ, Kanodia H, Falasconi A, Sigrist M, Esposito MS, Arber S. Ferreira-Pinto MJ, et al. Cell. 2021 Aug 19;184(17):4564-4578.e18. doi: 10.1016/j.cell.2021.07.002. Epub 2021 Jul 23. Cell. 2021. PMID: 34302739 Free PMC article. - The basal ganglia and the locomotor regions.
Garcia-Rill E. Garcia-Rill E. Brain Res. 1986 Mar;396(1):47-63. Brain Res. 1986. PMID: 2871904 Review. - Motor Control: Illuminating an Enigmatic Midbrain Locomotor Center.
Esposito MS, Arber S. Esposito MS, et al. Curr Biol. 2016 Apr 4;26(7):R291-3. doi: 10.1016/j.cub.2016.02.043. Curr Biol. 2016. PMID: 27046818
Cited by
- A Brainstem Locomotor Circuit Drives the Activity of Speed Cells in the Medial Entorhinal Cortex.
Carvalho MM, Tanke N, Kropff E, Witter MP, Moser MB, Moser EI. Carvalho MM, et al. Cell Rep. 2020 Sep 8;32(10):108123. doi: 10.1016/j.celrep.2020.108123. Cell Rep. 2020. PMID: 32905779 Free PMC article. - A brainstem monosynaptic excitatory pathway that drives locomotor activities and sympathetic cardiovascular responses.
Koba S, Kumada N, Narai E, Kataoka N, Nakamura K, Watanabe T. Koba S, et al. Nat Commun. 2022 Aug 29;13(1):5079. doi: 10.1038/s41467-022-32823-x. Nat Commun. 2022. PMID: 36038592 Free PMC article. - Mesencephalic locomotor region stimulation-cuneiform or pedunculopontine?
Burnside ER, Bradke F. Burnside ER, et al. Cell Rep Med. 2023 Feb 21;4(2):100948. doi: 10.1016/j.xcrm.2023.100948. Cell Rep Med. 2023. PMID: 36812884 Free PMC article. - Aberrant features of in vivo striatal dynamics in Parkinson's disease.
Lee K, Masmanidis SC. Lee K, et al. J Neurosci Res. 2019 Dec;97(12):1678-1688. doi: 10.1002/jnr.24519. Epub 2019 Sep 9. J Neurosci Res. 2019. PMID: 31502290 Free PMC article. Review. - Colocalized, bidirectional optogenetic modulations in freely behaving mice with a wireless dual-color optoelectronic probe.
Li L, Lu L, Ren Y, Tang G, Zhao Y, Cai X, Shi Z, Ding H, Liu C, Cheng D, Xie Y, Wang H, Fu X, Yin L, Luo M, Sheng X. Li L, et al. Nat Commun. 2022 Feb 11;13(1):839. doi: 10.1038/s41467-022-28539-7. Nat Commun. 2022. PMID: 35149715 Free PMC article.
References
- Albin RL, Young AB, Penney JB. The Functional-Anatomy of Basal Ganglia Disorders. Trends in Neurosciences. 1989;12:366–375. - PubMed
- Bateup HS, Santini E, Shen W, Birnbaum S, Valjent E, Surmeier DJ, Fisone G, Nestler EJ, Greengard P. Distinct subclasses of medium spiny neurons differentially regulate striatal motor behaviors. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:14845–14850. - PMC - PubMed
- Bedford TG, Loi PK, Crandall CC. A model of dynamic exercise: the decerebrate rat locomotor preparation. Journal of applied physiology. 1992;72:121–127. - PubMed
- Bjursten LM, Norrsell K, Norrsell U. Behavioural repertory of cats without cerebral cortex from infancy. Experimental brain research. 1976;25:115–130. - PubMed
- Borgius L, Restrepo CE, Leao RN, Saleh N, Kiehn O. A transgenic mouse line for molecular genetic analysis of excitatory glutamatergic neurons. Molecular and cellular neurosciences. 2010;45:245–257. - PubMed
Publication types
MeSH terms
Grants and funding
- P01 DA010154/DA/NIDA NIH HHS/United States
- R01 NS078435/NS/NINDS NIH HHS/United States
- T32 GM007618/GM/NIGMS NIH HHS/United States
- F31 NS092253/NS/NINDS NIH HHS/United States
- RR018928/RR/NCRR NIH HHS/United States
- Intramural NIH HHS/United States
- 5T32GM007618-38/GM/NIGMS NIH HHS/United States
- P30 DK057521/DK/NIDDK NIH HHS/United States
- R01 NS064984/NS/NINDS NIH HHS/United States
- C06 RR018928/RR/NCRR NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases