Resistant starch alters gut microbiome and metabolomic profiles concurrent with amelioration of chronic kidney disease in rats - PubMed (original) (raw)
Resistant starch alters gut microbiome and metabolomic profiles concurrent with amelioration of chronic kidney disease in rats
Dorothy A Kieffer et al. Am J Physiol Renal Physiol. 2016.
Abstract
Patients and animals with chronic kidney disease (CKD) exhibit profound alterations in the gut environment including shifts in microbial composition, increased fecal pH, and increased blood levels of gut microbe-derived metabolites (xenometabolites). The fermentable dietary fiber high amylose maize-resistant starch type 2 (HAMRS2) has been shown to alter the gut milieu and in CKD rat models leads to markedly improved kidney function. The aim of the present study was to identify specific cecal bacteria and cecal, blood, and urinary metabolites that associate with changes in kidney function to identify potential mechanisms involved with CKD amelioration in response to dietary resistant starch. Male Sprague-Dawley rats with adenine-induced CKD were fed a semipurified low-fiber diet or a high-fiber diet [59% (wt/wt) HAMRS2] for 3 wk (n = 9 rats/group). The cecal microbiome was characterized, and cecal contents, serum, and urine metabolites were analyzed. HAMRS2-fed rats displayed decreased cecal pH, decreased microbial diversity, and an increased Bacteroidetes-to-Firmicutes ratio. Several uremic retention solutes were altered in the cecal contents, serum, and urine, many of which had strong correlations with specific gut bacteria abundances, i.e., serum and urine indoxyl sulfate were reduced by 36% and 66%, respectively, in HAMRS2-fed rats and urine p-cresol was reduced by 47% in HAMRS2-fed rats. Outcomes from this study were coincident with improvements in kidney function indexes and amelioration of CKD outcomes previously reported for these rats, suggesting an important role for microbial-derived factors and gut microbe metabolism in regulating host kidney function.
Keywords: chronic kidney disease; dietary fiber; gut microbiota; resistant starch; uremic retention solutes.
Copyright © 2016 the American Physiological Society.
Figures
Fig. 1.
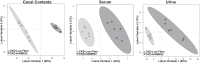
Principal coordinate analysis score plots of cecal contents, serum, and urine metabolites from male rats with chronic kidney disease (CKD) in the model validation group fed a low fiber diet or high amylose maize-resistant starch type 2 (HAMRS2). Ellipses represent 95% confidence intervals based on Hotelling's _T_2 statistic, and each symbol represents a rat. Metabolites that contributed to these plots are shown in Tables 2–4. Metabolomic analysis was performed on 9 rats/group, the model was developed using 6 rats/group, and model validation was performed using 3 rats/group.
Fig. 2.
A: unweighted UniFrac beta-diversity principal coordinate analysis plot displays separation between treatment groups based on the cecal microbiota of male CKD rats fed a low-fiber diet or HAMRS2. Axes represent percentage of the variance that can be accounted for based on the cecal microbiota profile. Ellipses represent 95% confidence intervals based on Hotelling's _T_2 statistic, and each symbol represents a rat. n = 9 rats/group. B: percent change of select cecal bacteria in CKD HAMRS2-fed rats relative to CKD low fiber-fed rats. Bacteria included had a minimum of 0.05% mean abundance in each group and P values of ≤0.05. Bacteria are listed to the lowest level of classification (i.e., if the last taxon assignment is f_, family is the lowest level of classification; p_ is phylum, o_ is order, and g_ is genus). n = 9 rats/group
Fig. 3.
Partial least squares-discriminant analysis scores plots based on serum, urine, and cecal metabolites of male CKD rats fed a low fiber diet or HAMRS2. Ellipses represent 95% confidence intervals based on Hotelling's _T_2 statistic, and each symbol represents a rat. Metabolites that contributed to these plots are shown in Tables 2–4. Metabolomic analysis was performed on 9 rats/group, the model was developed using 6 rats/group, and model validation was performed using 3 rats/group. QIPH, quantifier ion peak height.
Fig. 4.
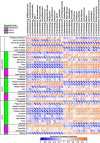
Spearman's correlation matrix of cecal bacteria versus metadata and uremic retention solutes in the cecal contents, serum, and urine of male CKD rats fed a low fiber diet or HAMRS2 (n = 18 total rats combined). Bacteria included had a minimum of 0.05% mean abundance in each group and an adjusted Mann-Whitney _U_-test P value of ≤0.05. Bacteria are listed to the lowest level of classification. Metabolites were selected based on being identified as uremic retention solutes. *Metabolites that had a mean bootstrapped variable importance in projection of ≥1. Colonic tight junction data were imputed for 2 rats/group using _k_-nearest neighbors, as described in
materials and methods
. The direction of ellipses represent positive or negative correlation, and the width of ellipse represents strength of correlation (narrow ellipse = stronger correlation).
Fig. 5.
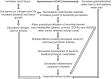
Working model of how HAMRS2 may improve CKD. SCFAs, short-chain fatty acids.
Similar articles
- Mice Fed a High-Fat Diet Supplemented with Resistant Starch Display Marked Shifts in the Liver Metabolome Concurrent with Altered Gut Bacteria.
Kieffer DA, Piccolo BD, Marco ML, Kim EB, Goodson ML, Keenan MJ, Dunn TN, Knudsen KE, Martin RJ, Adams SH. Kieffer DA, et al. J Nutr. 2016 Dec;146(12):2476-2490. doi: 10.3945/jn.116.238931. Epub 2016 Nov 2. J Nutr. 2016. PMID: 27807042 Free PMC article. - The Phosphate Binder Ferric Citrate Alters the Gut Microbiome in Rats with Chronic Kidney Disease.
Lau WL, Vaziri ND, Nunes ACF, Comeau AM, Langille MGI, England W, Khazaeli M, Suematsu Y, Phan J, Whiteson K. Lau WL, et al. J Pharmacol Exp Ther. 2018 Dec;367(3):452-460. doi: 10.1124/jpet.118.251389. Epub 2018 Oct 4. J Pharmacol Exp Ther. 2018. PMID: 30287477 - Modulation of the Gut Microbiota by Resistant Starch as a Treatment of Chronic Kidney Diseases: Evidence of Efficacy and Mechanistic Insights.
Snelson M, Kellow NJ, Coughlan MT. Snelson M, et al. Adv Nutr. 2019 Mar 1;10(2):303-320. doi: 10.1093/advances/nmy068. Adv Nutr. 2019. PMID: 30668615 Free PMC article. Review. - Metaproteomics reveals potential mechanisms by which dietary resistant starch supplementation attenuates chronic kidney disease progression in rats.
Zybailov BL, Glazko GV, Rahmatallah Y, Andreyev DS, McElroy T, Karaduta O, Byrum SD, Orr L, Tackett AJ, Mackintosh SG, Edmondson RD, Kieffer DA, Martin RJ, Adams SH, Vaziri ND, Arthur JM. Zybailov BL, et al. PLoS One. 2019 Jan 30;14(1):e0199274. doi: 10.1371/journal.pone.0199274. eCollection 2019. PLoS One. 2019. PMID: 30699108 Free PMC article. - The Impact of CKD on Uremic Toxins and Gut Microbiota.
Rysz J, Franczyk B, Ławiński J, Olszewski R, Ciałkowska-Rysz A, Gluba-Brzózka A. Rysz J, et al. Toxins (Basel). 2021 Mar 31;13(4):252. doi: 10.3390/toxins13040252. Toxins (Basel). 2021. PMID: 33807343 Free PMC article. Review.
Cited by
- Development and optimization of spray-dried functional oil microcapsules: Oxidation stability and release kinetics.
Yue H, Qiu B, Jia M, Liu J, Wang J, Huang F, Xu T. Yue H, et al. Food Sci Nutr. 2020 Jul 20;8(9):4730-4738. doi: 10.1002/fsn3.1684. eCollection 2020 Sep. Food Sci Nutr. 2020. PMID: 32994934 Free PMC article. - Exploring the Effects of Probiotic Treatment on Urinary and Serum Metabolic Profiles in Healthy Individuals.
Di Cesare F, Calgaro M, Ghini V, Squarzanti DF, De Prisco A, Visciglia A, Zanetta P, Rolla R, Savoia P, Amoruso A, Azzimonti B, Vitulo N, Tenori L, Luchinat C, Pane M. Di Cesare F, et al. J Proteome Res. 2023 Dec 1;22(12):3866-3878. doi: 10.1021/acs.jproteome.3c00548. Epub 2023 Nov 16. J Proteome Res. 2023. PMID: 37970754 Free PMC article. - The clinical impact of gut microbiota in chronic kidney disease.
Kim SM, Song IH. Kim SM, et al. Korean J Intern Med. 2020 Nov;35(6):1305-1316. doi: 10.3904/kjim.2020.411. Epub 2020 Sep 29. Korean J Intern Med. 2020. PMID: 32872729 Free PMC article. Review. - Gut microbial metabolites as mediators of renal disease: do short-chain fatty acids offer some hope?
Heaney LM, Davies OG, Selby NM. Heaney LM, et al. Future Sci OA. 2019 May 3;5(4):FSO384. doi: 10.4155/fsoa-2019-0013. Future Sci OA. 2019. PMID: 31114707 Free PMC article. No abstract available. - Gut microbiome in chronic kidney disease: challenges and opportunities.
Nallu A, Sharma S, Ramezani A, Muralidharan J, Raj D. Nallu A, et al. Transl Res. 2017 Jan;179:24-37. doi: 10.1016/j.trsl.2016.04.007. Epub 2016 Apr 30. Transl Res. 2017. PMID: 27187743 Free PMC article. Review.
References
- Annison G, Topping DL. Nutritional role of resistant starch: chemical structure vs physiological function. Annu Rev Nutr 14: 297–320, 1994. - PubMed
- Baker D. Determing fiber in cereals. Cereal Chem 54: 360–365, 1977.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical