Integrative approaches for large-scale transcriptome-wide association studies - PubMed (original) (raw)
doi: 10.1038/ng.3506. Epub 2016 Feb 8.
Alexander Gusev 1 2 3, Huwenbo Shi 6, Gaurav Bhatia 1 2 3, Wonil Chung 1, Brenda W J H Penninx 7, Rick Jansen 7, Eco J C de Geus 8, Dorret I Boomsma 8, Fred A Wright 9, Patrick F Sullivan 10 11 12, Elina Nikkola 4, Marcus Alvarez 4, Mete Civelek 13, Aldons J Lusis 4 13, Terho Lehtimäki 14, Emma Raitoharju 14, Mika Kähönen 15, Ilkka Seppälä 14, Olli T Raitakari 16 17, Johanna Kuusisto 18, Markku Laakso 18, Alkes L Price 1 2 3, Päivi Pajukanta 4 5, Bogdan Pasaniuc 4 6 19
Affiliations
- PMID: 26854917
- PMCID: PMC4767558
- DOI: 10.1038/ng.3506
Integrative approaches for large-scale transcriptome-wide association studies
Alexander Gusev et al. Nat Genet. 2016 Mar.
Abstract
Many genetic variants influence complex traits by modulating gene expression, thus altering the abundance of one or multiple proteins. Here we introduce a powerful strategy that integrates gene expression measurements with summary association statistics from large-scale genome-wide association studies (GWAS) to identify genes whose cis-regulated expression is associated with complex traits. We leverage expression imputation from genetic data to perform a transcriptome-wide association study (TWAS) to identify significant expression-trait associations. We applied our approaches to expression data from blood and adipose tissue measured in ∼ 3,000 individuals overall. We imputed gene expression into GWAS data from over 900,000 phenotype measurements to identify 69 new genes significantly associated with obesity-related traits (BMI, lipids and height). Many of these genes are associated with relevant phenotypes in the Hybrid Mouse Diversity Panel. Our results showcase the power of integrating genotype, gene expression and phenotype to gain insights into the genetic basis of complex traits.
Conflict of interest statement
The authors declare no competing financial interests.
Figures
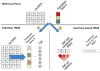
Figure 1. Overview of methods
Cartoon representation of TWAS approach. In the reference panel (top) estimate gene expression effect-sizes: directly (i.e. eQTL); modeling LD (BLUP); or modeling LD and effect-sizes (BSLMM). A: Predict expression directly into genotyped samples using effect-sizes from the reference panel and measure association between predicted expression and trait. B: Indirectly estimate association between predicted expression and trait as weighted linear combination of SNP-trait standardized effect sizes while accounting for LD among SNPs.
Figure 2. Modes of expression causality
Diagrams are shown for the possible modes of causality for the relationship between genetic markers (SNP, blue), gene expression (GE, green), and trait (red). A–D describes scenarios that would be considered null by the TWAS model; E–G describes scenarios that could be identified as significant.
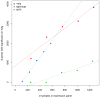
Figure 3. Number of genes with significant cis-heritability observed at varying sample sizes
The number of genes with significant cis-heritability was estimated by down-sampling each cohort (YFS, METSIM, and NTR/Wright et al.) into quintiles.
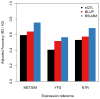
Figure 4. Accuracy of direct expression imputation algorithms
Adjusted accuracy was estimated using cross-validation R^2 between prediction and true expression, and normalized by corresponding cis-h2g. Bars show mean estimate across three cohorts and three methods: eQTL – single best cis-eQTL in the locus; BLUP using all SNPs in the locus; BSLMM using all SNPs in the locus and non-infinitesimal priors.
Figure 5. Power of summary-based expression imputation algorithms
Realistic disease architectures were simulated and power to detect a genome-wide significant association evaluated across three methods (accounting for 15,000 eGWAS/TWAS tests, and 1,000,000 GWAS tests). Colors correspond number of causal variants simulated and methods used: GWAS where every SNP in the locus is tested; eGWAS where only the best cis-eQTL is tested; and TWAS computed using summary-statistics. Expression reference panel was fixed at 1,000 out-of-sample individuals and simulated GWAS sample size designated by x-axis. Power was computed as the fraction of 500 simulations where significant association was identified.
Comment in
- Complex traits: Integrating gene variation and expression to understand complex traits.
Cloney R. Cloney R. Nat Rev Genet. 2016 Apr;17(4):194. doi: 10.1038/nrg.2016.18. Epub 2016 Feb 22. Nat Rev Genet. 2016. PMID: 26900024 No abstract available.
Similar articles
- Leveraging expression from multiple tissues using sparse canonical correlation analysis and aggregate tests improves the power of transcriptome-wide association studies.
Feng H, Mancuso N, Gusev A, Majumdar A, Major M, Pasaniuc B, Kraft P. Feng H, et al. PLoS Genet. 2021 Apr 8;17(4):e1008973. doi: 10.1371/journal.pgen.1008973. eCollection 2021 Apr. PLoS Genet. 2021. PMID: 33831007 Free PMC article. - Learning from Fifteen Years of Genome-Wide Association Studies in Age-Related Macular Degeneration.
Strunz T, Kiel C, Sauerbeck BL, Weber BHF. Strunz T, et al. Cells. 2020 Oct 10;9(10):2267. doi: 10.3390/cells9102267. Cells. 2020. PMID: 33050425 Free PMC article. Review. - How powerful are summary-based methods for identifying expression-trait associations under different genetic architectures?
Veturi Y, Ritchie MD. Veturi Y, et al. Pac Symp Biocomput. 2018;23:228-239. Pac Symp Biocomput. 2018. PMID: 29218884 Free PMC article. - Aggregating multiple expression prediction models improves the power of transcriptome-wide association studies.
Zeng P, Dai J, Jin S, Zhou X. Zeng P, et al. Hum Mol Genet. 2021 May 29;30(10):939-951. doi: 10.1093/hmg/ddab056. Hum Mol Genet. 2021. PMID: 33615361 - Opportunities and challenges for transcriptome-wide association studies.
Wainberg M, Sinnott-Armstrong N, Mancuso N, Barbeira AN, Knowles DA, Golan D, Ermel R, Ruusalepp A, Quertermous T, Hao K, Björkegren JLM, Im HK, Pasaniuc B, Rivas MA, Kundaje A. Wainberg M, et al. Nat Genet. 2019 Apr;51(4):592-599. doi: 10.1038/s41588-019-0385-z. Epub 2019 Mar 29. Nat Genet. 2019. PMID: 30926968 Free PMC article. Review.
Cited by
- Leveraging expression from multiple tissues using sparse canonical correlation analysis and aggregate tests improves the power of transcriptome-wide association studies.
Feng H, Mancuso N, Gusev A, Majumdar A, Major M, Pasaniuc B, Kraft P. Feng H, et al. PLoS Genet. 2021 Apr 8;17(4):e1008973. doi: 10.1371/journal.pgen.1008973. eCollection 2021 Apr. PLoS Genet. 2021. PMID: 33831007 Free PMC article. - Challenges and novel approaches for investigating molecular mediation.
Richmond RC, Hemani G, Tilling K, Davey Smith G, Relton CL. Richmond RC, et al. Hum Mol Genet. 2016 Oct 1;25(R2):R149-R156. doi: 10.1093/hmg/ddw197. Epub 2016 Jul 20. Hum Mol Genet. 2016. PMID: 27439390 Free PMC article. - Learning from Fifteen Years of Genome-Wide Association Studies in Age-Related Macular Degeneration.
Strunz T, Kiel C, Sauerbeck BL, Weber BHF. Strunz T, et al. Cells. 2020 Oct 10;9(10):2267. doi: 10.3390/cells9102267. Cells. 2020. PMID: 33050425 Free PMC article. Review. - Brain proteome-wide association study implicates novel proteins in depression pathogenesis.
Wingo TS, Liu Y, Gerasimov ES, Gockley J, Logsdon BA, Duong DM, Dammer EB, Lori A, Kim PJ, Ressler KJ, Beach TG, Reiman EM, Epstein MP, De Jager PL, Lah JJ, Bennett DA, Seyfried NT, Levey AI, Wingo AP. Wingo TS, et al. Nat Neurosci. 2021 Jun;24(6):810-817. doi: 10.1038/s41593-021-00832-6. Epub 2021 Apr 12. Nat Neurosci. 2021. PMID: 33846625 Free PMC article. - Making Biological Sense of Genetic Studies of Age-Related Macular Degeneration.
Singh N, Swaroop A, Ratnapriya R. Singh N, et al. Adv Exp Med Biol. 2021;1256:201-219. doi: 10.1007/978-3-030-66014-7_8. Adv Exp Med Biol. 2021. PMID: 33848003
References
Publication types
MeSH terms
Grants and funding
- F31 HL127921/HL/NHLBI NIH HHS/United States
- F32 GM106584/GM/NIGMS NIH HHS/United States
- T32 HG002536/HG/NHGRI NIH HHS/United States
- P01 HL028481/HL/NHLBI NIH HHS/United States
- R25 GM055052/GM/NIGMS NIH HHS/United States
- T32HG002536/HG/NHGRI NIH HHS/United States
- HL-095056/HL/NHLBI NIH HHS/United States
- R01 GM053725/GM/NIGMS NIH HHS/United States
- HL-28481/HL/NHLBI NIH HHS/United States
- R01 GM105857/GM/NIGMS NIH HHS/United States
- R01 HL095056/HL/NHLBI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases