Developing genetic tools to exploit Chaetomium thermophilum for biochemical analyses of eukaryotic macromolecular assemblies - PubMed (original) (raw)
Developing genetic tools to exploit Chaetomium thermophilum for biochemical analyses of eukaryotic macromolecular assemblies
Nikola Kellner et al. Sci Rep. 2016.
Abstract
We describe a method to genetically manipulate Chaetomium thermophilum, a eukaryotic thermophile, along with various biochemical applications. The transformation method depends on a thermostable endogenous selection marker operating at high temperatures combined with chromosomal integration of target genes. Our technique allows exploiting eukaryotic thermophiles as source for purifying thermostable native macromolecular complexes with an emphasis on the nuclear pore complex, holding great potential for applications in basic science and biotechnology.
Figures
Figure 1. Schematic representation of the workflow for transformation of C. thermophilum protoplasts and selection of positive transformants.
Protoplasts are generated from C. thermophilum mycelium growing in submerged cultures by digestion of fungal cell walls. Upon filtration, protoplasts are plated on osmotically stabilizing medium to assess viability. For transformation, protoplasts are mixed with the P ACTIN -ERG1 plasmid for selection on terbinafine (see text for details). Arrowheads point to restriction sites for linearization. Upon recovery, mycelia are plated on solid media supplemented with terbinafine for selection of positive strains. Re-plating of individual transformed mycelia on selective medium confirms stable terbinafine resistance, whereas the wildtype does not grow on terbinafine. Left panel: PCR analysis with oligonucleotides specific for the P ACTIN -ERG1 construct (depicted as red and blue arrows, respectively) performed on genomic DNA (gDNA) isolated from five independent ERG1 transformants (lanes 1–5) to verify genomic integration of the construct. The expected products of 761 nt (upper panel) and 536 nt (lower panel), respectively, could be amplified, whereas no product was amplified from wildtype gDNA (wt). The P ACTIN -ERG1 plasmid (P) was included as positive control (lane 7). Ascospores (bottom left) were generated from an _ERG1_-positive transformant to verify that the resistance trait remains stable. Upon plating of these ascospores on terbinafine, the _ERG1_- positive strain was able to germinate, whereas ascospores generated from the wildtype strain did not grow. Bottom right: PCR analysis performed on gDNA isolated from four independent ascospore-derived clones (lanes 1–4) verifies stable genomic integration of the _ERG1_-marker.
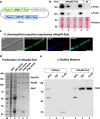
Figure 2. Design and expression of constructs for affinity purification of native C. thermophilum proteins.
(a) Schematic depiction of the plasmid used for transformation and integration of a C-terminally ProtA-Flag tagged _ct_Nup82 protein. (b) Western analysis of four strains positive for the P ACTIN -ERG1_P NUP82 -NUP82-FpA construct. Expression of the fusion protein was revealed with anti-ProteinA and anti-Flag antibodies on whole cell protein lysates, respectively, whereas no signal was detectable in the wildtype. Equal loading was confirmed by Ponceau S staining. A molecular weight standard with molecular weights (kDa) is depicted on the left. (c) Indirect immunofluorescence to detect C. thermophilum nuclear pore complexes. An antibody directed against Protein A in combination with an AlexaFluor488-conjugated secondary antibody decorates nuclear rims in the strain expressing _ct_Nup82-FpA, indicating that the fusion protein is functional and localizes to the NPC. DAPI staining was performed to visualize nuclei. DIC, GFP and DAPI channels as well as merged fluorescent images are shown. Scale bars – 10 μm. (d) Tandem-affinity purification of the native _ct_Nup82 complex. Samples are analyzed by SDS-PAGE and Coomassie staining. Whole cell lysate (WCL), the complex on IgG- and Flag-beads, respectively, the TEV- and final Flag-eluates are shown. Proteins were identified by mass-spectrometry. (e) Western analysis to identify O-linked glycosylation. Wildtype and _ct_Nup82-FpA expressing mycelium was subjected to affinity purification. WCL as well as TEV and Flag eluates, respectively, were probed with an antibody specific for O-linked N-acetylglucosamines (O-GlcNAc). This antibody detects a band with a molecular weight corresponding to _ct_Nsp1, both in the whole cell lysate and the purified trimeric complex, indicating O-GlcNAc modification of this nucleoporin. A molecular weight standard with molecular weights (kDa) is depicted on the left.
Figure 3. Biochemistry of native C. thermophilum nuclear pore subcomplexes and reconstitution of a Nup82-Nup84 super-complex.
(a) Affinity capture of _ct_Nup53-FpA was performed on cleared lysates under various extraction conditions. Elution of the complexes was followed by SDS-PAGE and Coomassie staining. Proteins identified by mass spectrometric analysis from the complexes isolated in condition #9 are indicated. Extraction conditions (# 1–14) are presented in Supplementary Table 1. A molecular weight marker (M) is indicated on the left. (b) Sucrose gradient centrifugation of the _ct_Nup82 complex upon affinity capture of _ct_Nup82-FpA. Fractions were collected and analyzed by SDS-PAGE and Coomassie staining. Individual proteins were identified by mass-spectrometric analysis. (c) In vitro binding assay with an immobilized Nup82•Nup159C1•Nsp1C complex (_ct_Nup82 complex) and full length _ct_Nup120, _ct_Nup85 and _ct_Nup145C (_ct_Nup84 complex) (input, lanes 1–3). Samples are eluted via Flag-Peptide and analyzed by SDS-PAGE and Coomassie staining (lanes 5–11). MW: molecular weight, kDa: Kilodalton, α-Flag: Anti-Flag M2 Affinity Gel. (d) Glycerol-gradient centrifugation of in vitro reconstituted complexes. Fractions were collected and analyzed by SDS-PAGE and Coomassie staining. (top) _ct_Nup82 complex, (middle) _ct_Nup84 complex including _ct_Nup84, (bottom) _ct_Nup82-_ct_Nup84 complex including _ct_Nup84. (e) Schematic model for the connection of the Nup53-containing inner pore ring complex (IRC) with the Nup82 outer ring and the Nsp1 channel complex (left) and how the Nup82 complex might be linked to the Y-shaped octameric Nup84 complex within the NPC protomer.
Similar articles
- Transformation of Chaetomium thermophilum and Affinity Purification of Native Thermostable Protein Complexes.
Kellner N, Hurt E. Kellner N, et al. Methods Mol Biol. 2022;2502:35-50. doi: 10.1007/978-1-0716-2337-4_2. Methods Mol Biol. 2022. PMID: 35412229 - A Homologous Recombination System to Generate Epitope-Tagged Target Genes in Chaetomium thermophilum: A Genetic Approach to Investigate Native Thermostable Proteins.
Kellner N, Griesel S, Hurt E. Kellner N, et al. Int J Mol Sci. 2022 Mar 16;23(6):3198. doi: 10.3390/ijms23063198. Int J Mol Sci. 2022. PMID: 35328616 Free PMC article. - Insight into structure and assembly of the nuclear pore complex by utilizing the genome of a eukaryotic thermophile.
Amlacher S, Sarges P, Flemming D, van Noort V, Kunze R, Devos DP, Arumugam M, Bork P, Hurt E. Amlacher S, et al. Cell. 2011 Jul 22;146(2):277-89. doi: 10.1016/j.cell.2011.06.039. Cell. 2011. PMID: 21784248 - The nuclear pore complex.
Adam SA. Adam SA. Genome Biol. 2001;2(9):REVIEWS0007. doi: 10.1186/gb-2001-2-9-reviews0007. Epub 2001 Aug 31. Genome Biol. 2001. PMID: 11574060 Free PMC article. Review. - [Structural organization, functions and dynamics of nuclear pores].
Gubanova NV, Morozova KN, Kiseleva EV. Gubanova NV, et al. Tsitologiia. 2006;48(11):887-99. Tsitologiia. 2006. PMID: 17233474 Review. Russian.
Cited by
- Interaction network of the ribosome assembly machinery from a eukaryotic thermophile.
Baßler J, Ahmed YL, Kallas M, Kornprobst M, Calviño FR, Gnädig M, Thoms M, Stier G, Ismail S, Kharde S, Castillo N, Griesel S, Bastuck S, Bradatsch B, Thomson E, Flemming D, Sinning I, Hurt E. Baßler J, et al. Protein Sci. 2017 Feb;26(2):327-342. doi: 10.1002/pro.3085. Epub 2017 Jan 14. Protein Sci. 2017. PMID: 27863450 Free PMC article. - Protoplast-mediated transformation of Madurella mycetomatis using hygromycin resistance as a selection marker.
du Pré S, Konings M, Schoorl DJA, Fahal AH, Arentshorst M, Ram AFJ, van de Sande WWJ. du Pré S, et al. PLoS Negl Trop Dis. 2024 Apr 5;18(4):e0012092. doi: 10.1371/journal.pntd.0012092. eCollection 2024 Apr. PLoS Negl Trop Dis. 2024. PMID: 38578808 Free PMC article. - Mechanism of 5S RNP recruitment and helicase-surveilled rRNA maturation during pre-60S biogenesis.
Lau B, Huang Z, Kellner N, Niu S, Berninghausen O, Beckmann R, Hurt E, Cheng J. Lau B, et al. EMBO Rep. 2023 Jul 5;24(7):e56910. doi: 10.15252/embr.202356910. Epub 2023 May 2. EMBO Rep. 2023. PMID: 37129998 Free PMC article. - Polysaccharide monooxygenase-catalyzed oxidation of cellulose to glucuronic acid-containing cello-oligosaccharides.
Chen J, Guo X, Zhu M, Chen C, Li D. Chen J, et al. Biotechnol Biofuels. 2019 Feb 27;12:42. doi: 10.1186/s13068-019-1384-0. eCollection 2019. Biotechnol Biofuels. 2019. PMID: 30858879 Free PMC article. - Ribosome origami.
Rorbach J, Aibara S, Amunts A. Rorbach J, et al. Nat Struct Mol Biol. 2017 Nov 7;24(11):879-881. doi: 10.1038/nsmb.3497. Nat Struct Mol Biol. 2017. PMID: 29112687 No abstract available.
References
- Elleuche S., Schafers C., Blank S., Schroder C. & Antranikian G. Exploration of extremophiles for high temperature biotechnological processes. Current opinion in microbiology 25, 113–119 (2015). - PubMed
- Cava F., Hidalgo A. & Berenguer J. Thermus thermophilus as biological model. Extremophiles: life under extreme conditions 13, 213–231 (2009). - PubMed
- Amlacher S. et al.. Insight into structure and assembly of the nuclear pore complex by utilizing the genome of a eukaryotic thermophile. Cell 146, 277–289 (2011). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources